User login
Administration of initially appropriate antimicrobial therapy represents a key determinant of outcome in patients with severe infection.[1, 2, 3, 4, 5, 6, 7, 8, 9] The variable patterns of antimicrobial resistance seen between and within healthcare institutions complicate the process of antibiotic selection. Although much attention has historically focused on Staphylococcus aureus, resistance among Gram‐negative pathogens has emerged as a major challenge in the care of hospitalized, and particularly critically ill, patients.[2, 10, 11] Multidrug, and more specifically carbapenem resistance, among such common organisms as Pseudomonas aeruginosa (PA) and Enterobacteriaceae represents a major treatment challenge.[2] A recent US‐based surveillance study reported that a quarter of device‐related infections in hospitalized patients were caused by carbapenem‐resistant PA.[10]
In addition to changes in resistance patterns seen among PA isolates, increasing rates of nonsusceptibility have been described among Enterobacteriaceae. Resistance rates to third‐generation cephalosporins in these pathogens have risen steadily since 1988, reaching 20% among Klebsiella pneumoniae and 5% among Escherichia coli isolates by 2004.[11] In response to this, clinicians have increasingly utilized carbapenems to treat patients with serious Gram‐negative infections. However, the development of several types of carbapenemases by Enterobacteriaceae has led to a greater prevalence of carbapenem‐resistant Enterobacteriaceae species (CRE).[12, 13, 14, 15, 16, 17, 18] In fact, a recent report from the Centers for Disease Control and Prevention (CDC) documents a rapid rise in both the prevalence and extent of CRE in the United States.[19]
These Gram‐negative multidrug‐resistant (MDR) organisms frequently cause serious infections including pneumonia and bloodstream infections (BSI). The fact that these conditions, if not addressed in a timely and appropriate manner, lead to high morbidity, mortality, and costs, makes understanding the patterns of resistance that much more critical. To gain a better understanding of the prevalence and characteristics of MDR rates among PA and carbapenem resistance in Enterobacteriaceae in patients hospitalized in the United States with pneumonia and BSI, we conducted a multicenter survey of microbiology data.
METHODS
To determine the prevalence of predefined resistance patterns among PA and Enterobacteriaceae in pneumonia and BSI specimens, we examined The Surveillance Network (TSN) database from Eurofins between years 2000 and 2009. The database has been used extensively for surveillance purposes since 1994 and has previously been described in detail.[17, 20, 21, 22, 23] Briefly, TSN is a warehouse of routine clinical microbiology data collected from a nationally representative sample of microbiology laboratories in 217 hospitals in the United States. To minimize selection bias, laboratories are included based on their geography and the demographics of the populations they serve.[20] Only clinically significant samples are reported. No personal identifying information for source patients is available in this database. Only source laboratories that perform antimicrobial susceptibility testing according standard US Food and Drug Administration‐approved testing methods and interpret susceptibility in accordance with the Clinical Laboratory Standards Institute breakpoints are included.[24] All enrolled laboratories undergo a pre‐enrollment site visit. Logical filters are used for routine quality control to detect unusual susceptibility profiles and to ensure appropriate testing methods. Repeat testing and reporting are done as necessary.[20]
Laboratory samples are reported as susceptible, intermediate, or resistant.[24] We required that samples have susceptibility data for each of the antimicrobials needed to determine their resistance phenotype. These susceptibility patterns served as phenotypic surrogates for resistance. We grouped intermediate samples together with the resistant ones for the purposes of the current analysis. Duplicate isolates were excluded. Only samples representing 1 of the 2 infections of interest, pneumonia and BSI, were included.
We defined MDR‐PA as any isolate resistant to 3 of the following drug classes: aminoglycoside (gentamicin), antipseudomonal penicillin (piperacillin‐tazobactam), antipseudomonal cephalosporin (ceftazidime), carbapenems (imipenem, meropenem), and fluoroquinolone (ciprofloxacin). Enterobacteriaceae were considered CRE if resistant to both a third‐generation cephalosporin and a carbapenem. We examined the data by infection type, year, the 9 US Census geographical divisions, and intensive care unit (ICU) origin.
We did not pursue hypothesis testing due to a high risk of type I error in this large dataset. Therefore, only clinically important trends are highlighted.
RESULTS
Source specimen characteristics for the 205,526 PA (187,343 pneumonia and 18,183 BSI) and 95,566 Enterobacteriaceae specimens (58,810 pneumonia and 36,756 BSI) identified are presented in Table 1. The median age of the patients from which the isolates derive was similar among the PA pneumonia, Enterobacteriaceae pneumonia, and Enterobacteriaceae BSI groups, but higher in the PA BSI group. Similarly, there were differences in the gender distribution of source patients between the organisms and infections. Namely, although females represented a stable 42% of each of the infections with PA, the proportions of females with Enterobacteriaceae pneumonia (36.2%) differed from that in the BSI group (48.6%). Pneumonia specimens (34.0% PA and 39.0% Enterobacteriaceae) were more likely to originate in the ICU than those from BSI (28.4% PA and 21.1% Enterobacteriaceae).
| Pseudomonas aeruginosa, N=205,526 | Enterobacteriaceae, N=95,566 | |
|---|---|---|
| ||
| Pneumonia, n | 187,343 | 58,810 |
| Age, y, median (IQR 25, 75) | 54 (23, 71) | 55 (21, 71) |
| Gender, female, n (%) | 78,418 (41.9) | 21,305 (36.2) |
| ICU origin, n (%) | 63,755 (34.0) | 22,942 (39.0) |
| Meeting definitions of resistance, n (%) | 41,180 (22.0) | 930 (1.6) |
| BSI, n | 18,183 | 36,756 |
| Age, y, median (IQR 25, 75) | 59 (31, 75) | 55 (24, 71) |
| Gender, female, n (%) | 7,448 (41.8) | 17,871 (48.6) |
| ICU origin, n (%) | 5,170 (28.4) | 7,751 (21.1) |
| Meeting definitions of resistance, n (%) | 2,668 (14.7) | 394 (1.1) |
The prevalence of resistance among PA isolates was approximately 15‐fold higher than among Enterobacteriaceae specimens in both infection types (Table 1). This pattern persisted when stratified by infection type (pneumonia: 22.0% MDR‐PA vs 1.6% CRE; BSI: 14.7% MDR‐PA vs 1.1% CRE).
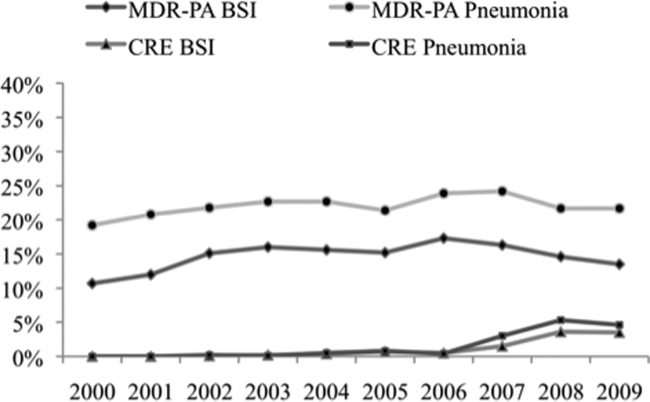
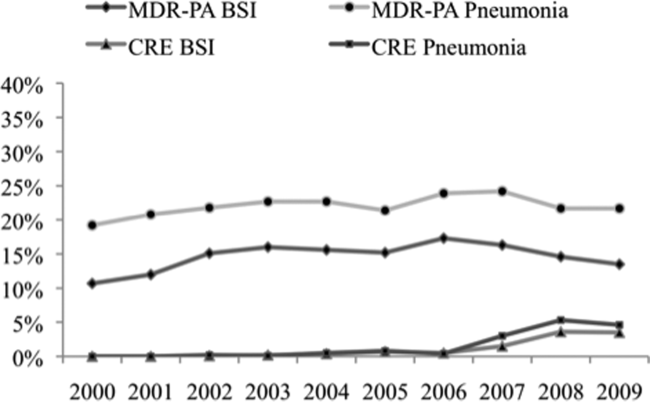
Over the time frame of the study, we detected variable patterns of resistance in the 2 groups of organisms (Figure 1). Namely, among PA in both pneumonia and BSI there was an initial rise in the proportion of MDR specimens between 2000 and 2003, followed by a stabilization until 2005, an additional rise in 2006, and a gradual decline and stabilization through 2009. These fluctuations notwithstanding, there was a net rise in MDR‐PA as a proportion of all PA from 10.7% in 2000 to 13.5% in 2009 among BSI, and from 19.2% in 2000 to 21.7% in 2009 among pneumonia specimens. Among Enterobacteriaceae, the CRE phenotype emerged in 2002 in both infection types and peaked in 2008 at 3.6% in BSI and 5.3% in pneumonia. This peak was followed by a stabilization in 2009 in BSI (3.5%) and a further decline, albeit minor, to 4.6% in pneumonia.
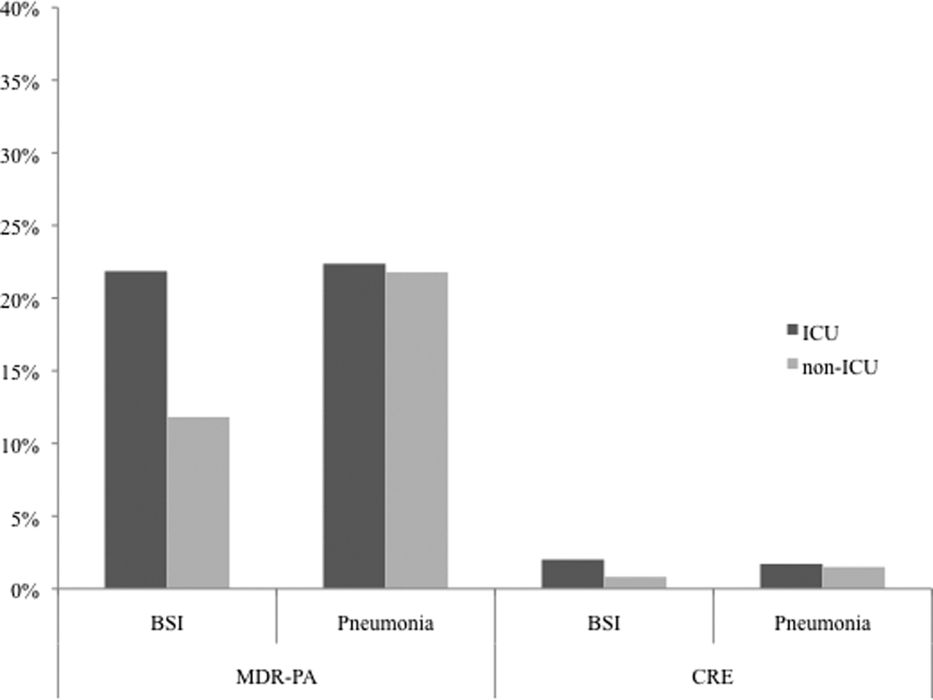
We noted geographic differences in the distribution of resistance (Table 2). Although MDR‐PA was more likely to originate from the East and West North Central divisions, and least likely from the New England and Mountain states, most CRE was detected in the specimens from the latter 2 regions. When stratified by ICU as the location of specimen origin, there were differences in the prevalence of resistant organisms of both types, but these differences were observed only in BSI specimens and not in pneumonia (Figure 2). That is, in BSI, the likelihood of a resistant organism originating from the ICU was approximately double that from a non‐ICU location for both MDR‐PA (21.9% vs 11.8%) and CRE (2.0% vs 0.8%).
| Census Division | MDR‐PA | CRE | ||
|---|---|---|---|---|
| BSI | Pneumonia | BSI | Pneumonia | |
| ||||
| East North Central | 20.8% | 26.9% | 2.0% | 1.9% |
| West North Central | 18.0% | 22.1% | 0.8% | 0.7% |
| East South Central | 15.8% | 20.5% | 0.1% | 0.1% |
| West South Central | 13.5% | 21.7% | 0.3% | 0.5% |
| Pacific | 13.1% | 20.3% | 0.3% | 0.3% |
| Mid‐Atlantic | 12.6% | 20.5% | 2.5% | 3.8% |
| South Atlantic | 12.6% | 21.6% | 0.9% | 1.5% |
| New England | 10.7% | 19.7% | 1.3% | 2.9% |
| Mountain | 8.5% | 19.4% | 0.4% | 1.1% |
DISCUSSION
We have demonstrated that among both pneumonia and BSI specimens, PA and Enterobacteriaceae have a high prevalence of multidrug resistance. When examined cross‐sectionally, in both pneumonia and BSI, the prevalence of MDR‐PA was approximately 15‐fold higher than the prevalence of CRE among Enterobacteriaceae. Over the time frame of the study, MDR‐PA rose and then fell and stabilized to levels only slightly higher than those observed at the beginning of the observation period. In contrast, CRE emerged and rose precipitously between 2006 and 2008, and appeared to stabilize in 2009 in both infection types. Interestingly, we observed geographic variability among resistant isolates. Specifically, the prevalence of CRE was highest in the region with a relatively low prevalence of MDR‐PA. Despite this heterogeneity geographically, resistance for both isolate types in BSI but not in pneumonia was substantially higher in the ICU than outside the ICU.
Our data enhance the current understanding of distribution of Gram‐negative resistance in the United States. A recent study by Braykov and colleagues examined time trends in the development of CRE phenotype among Klebsiella pneumoniae in the United States.[17] By focusing on this single pathogen in various infections within Eurofin's TSN database between 1999 and 2010, they pinpointed its initial emergence to year 2002, with a notably steep rise between 2006 and 2009, with some reduction in the pace of growth in 2010. We have documented an analogous rise in the CRE phenotype among all Enterobacteriaceae, particularly in pneumonia and BSI within a similar time period. Thus, our data on the 1 hand broaden the concern about this pathogen beyond just a single organism within Enterobacteraceae and a single antimicrobial class, and on the other hand serve to focus attention on 2 clinically burdensome infection types, pneumonia and BSI.
Another recent investigation reported a rise in carbapenem‐resistant Enterobacteriaceae in US hospitals over the past decade.[19] Drawing on data from multiple sources, including the dataset used for the current analysis, this study examined the patterns of single‐class resistance to carbapenems among central line‐associated BSI (CLABSI) and catheter‐associated urinary tract infection specimens. Consistent with our findings, these authors noted that the highest percentage of hospitals reporting such single‐class carbapenem‐resistant specimens were located in the Northeastern United States. They also described that the proportion of Enterobacteriaceae with single‐class carbapenem resistance rose from 0% in 2001 to 1.4% in 2010. An additional CDC analysis reported that single‐class carbapenem resistance now exists in 4.2% of Enterobacteraciae as compared to 1.2% of isolates in 2001. We confirm that this rise in single‐class resistance is echoed by a rise in the prevalence of the CRE phenotype, and provides further granularity to this problem, specifically in the setting of pneumonia and BSI.
Although CRE has become an important concern in the treatment of patients with pneumonia and BSI, MDR‐PA remains a far larger challenge in these infections. CREs appear to occur more frequently than in the past but remain relatively dwarfed by the prevalence of MDR‐PA. Our data are generally in agreement with the 2009 to 2010 data from the National Healthcare Safety Network (NHSN) maintained by the CDC, which focuses on CLABSI and ventilator‐associated pneumonia (VAP) rather than general BSI and pneumonia in US hospitals.[25] In this report, the proportion of PA that were classified as MDR according to a definition similar to ours was 15.4% in CLABSI and 17.7% in VAP. In contrast, we document that 13.5% of PA causing BSI and 21.7% causing pneumonia were due to MDR‐PA organisms. This mild divergence likely reflects the slightly different antimicrobials utilized to define MDR‐PA in the 2 studies, as well as variance in the populations examined. An additional data point reported in the NHSN study is the proportion of MDR‐PA CLABSI originating in the ICU (16.8%) versus non‐ICU hospital locations (13.3%). Although the difference we found in the prevalence of BSI by the location in the hospital was greater, we confirm that ICU specimens carry a higher risk of harboring MDR‐PA.
Our study has a number of strengths and limitations. Because we used a nationally representative database to derive our estimates, our results are highly generalizable.
The TSN database consists of microbiology samples from hospital laboratories. Although we attempted to reduce the risk of duplication, because of how samples are numbered in the database, repeat sampling remains a possibility. The definitions of resistance were based on phenotypic patterns of resistance to various antimicrobial classes. This makes our resistant organisms subject to misclassification.
In summary, although carbapenem resistance among Enterobacteriaceae has emerged as an important phenomenon, multidrug resistance among PA remains relatively more prevalent in the United States. Furthermore, over the decade examined, MDR‐PA has remained an important pathogen in pneumonia and BSI that persists across all geographic regions of the United States. Although CRE is rightfully receiving a disproportionate share of attention from public health officials, it would be shortsighted to ignore the importance of MDR‐PA as a target, not only for transmission prevention and antimicrobial stewardship, but also for new therapeutic development. Because the patterns of resistance are rapidly evolving, it is incumbent upon our public health enterprise to perform more granular real‐time surveillance to allow changes in epidemiology to inform policy and treatment decisions.
ACKNOWLEDGEMENTS
Disclosures: This study was supported by a grant from Cubist Pharmaceuticals. The authors report no conflicts of interest.
- National Nosocomial Infections Surveillance (NNIS) System Report. Am J Infect Control. 2004;32:470.
- , , , . National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob Agents Chemother. 2004;48:4606–4610.
- , , , et al. Health care‐associated pneumonia and community‐acquired pneumonia: a single‐center experience. Antimicrob Agents Chemother. 2007;51:3568–3573.
- , , , et al. Clinical importance of delays in the initiation of appropriate antibiotic treatment for ventilator‐associated pneumonia. Chest. 2002;122:262–268.
- . Modification of empiric antibiotic treatment in patients with pneumonia acquired in the intensive care unit. ICU‐Acquired Pneumonia Study Group. Intensive Care Med. 1996;22:387–394.
- , , , , . Antimicrobial therapy escalation and hospital mortality among patients with HCAP: a single center experience. Chest. 2008:134:963–968.
- , , , et al. Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock: 2008. Crit Care Med. 2008;36:296–327.
- , , , , , . Inappropriate antibiotic therapy in Gram‐negative sepsis increases hospital length of stay. Crit Care Med. 2011;39:46–51.
- , , , . Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients. Chest. 1999;115:462–474.
- , , , et al. Antimicrobial‐resistant pathogens associated with healthcare‐associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hospital Epidemiol. 2008;29:996–1011.
- , ; National Nosocomial Infections Surveillance (NNIS) System. Overview of nosocomial infections caused by Gram‐negative bacilli. Clin Infect Dis. 2005;41:848–854.
- , , . The real threat of Klebsiella pneumoniae carbapenemase‐producing bacteria. Lancet Infect Dis. 2009;9:228–236.
- , , , . Household transmission of carbapenemase‐producing Klebsiella pneumoniae. Emerg Infect Dis. 2008;14:859–860.
- , , , . Isolation of imipenem‐resistant Enterobacter species: emergence of KPC‐2 carbapenemase, molecular characterization, epidemiology, and outcomes. Antimicrob Agents Chemother. 2008;52:1413–1418.
- , , , , . Outcomes of carbapenem‐resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008;29:1099–1106.
- , , , , , ; for the Centers for Disease Control and Prevention Epicenter Program. Emergence and rapid regional spread of Klebsiella pneumoniae carbapenemase‐producing Enterobacteriaceae. Clin Infect Dis. 2011;53:532–540.
- , , , , . Trends in resistance to carbapenems and third‐generation cephalosporins among clinical isolates of Klebsiella pneumoniae in the United States, 1999–2010. Infect Control Hosp Epidemiol. 2013;34:259–268.
- , , , . Population‐based incidence of carbapenem‐resistant Klebsiella pneumoniae along the continuum of care, Los Angeles County. Infect Control Hosp Epidemiol. 2013;34:144–150.
- Centers for Disease Control and Prevention (CDC). Vital signs: carbapenem‐resistant enterobacteriaceae. MMWR Morb Mortal Wkly Rep. 2013;62:165–170.
- , , . Antimicrobial resistance in key bloodstream bacterial isolates: electronic surveillance with the Surveillance Network Database–USA. Clin Infect Dis. 1999;29:259–263.
- , , . Community‐associated methicillin‐resistant Staphylococcus aureus in outpatients, United States, 1999–2006. Emerg Infect Dis. 2009;15:1925–1930.
- , , . Increasing resistance of Acinetobacter species to imipenem in United States hospitals, 1999–2006. Infect Control Hosp Epidemiol. 2010;31:196–197.
- , , , , . Prevalence of antimicrobial resistance in bacteria isolated from central nervous system specimens as reported by U.S. hospital laboratories from 2000 to 2002. Ann Clin Microbiol Antimicrob. 2004;3:3.
- Clinical Laboratory Standards Institute. Available at: http://www.clsi.org. Accessed July 8, 2013.
- , , , et al. Antimicrobial‐resistant pathogens associates with healthcare‐associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol. 2013;34:1–14.
Administration of initially appropriate antimicrobial therapy represents a key determinant of outcome in patients with severe infection.[1, 2, 3, 4, 5, 6, 7, 8, 9] The variable patterns of antimicrobial resistance seen between and within healthcare institutions complicate the process of antibiotic selection. Although much attention has historically focused on Staphylococcus aureus, resistance among Gram‐negative pathogens has emerged as a major challenge in the care of hospitalized, and particularly critically ill, patients.[2, 10, 11] Multidrug, and more specifically carbapenem resistance, among such common organisms as Pseudomonas aeruginosa (PA) and Enterobacteriaceae represents a major treatment challenge.[2] A recent US‐based surveillance study reported that a quarter of device‐related infections in hospitalized patients were caused by carbapenem‐resistant PA.[10]
In addition to changes in resistance patterns seen among PA isolates, increasing rates of nonsusceptibility have been described among Enterobacteriaceae. Resistance rates to third‐generation cephalosporins in these pathogens have risen steadily since 1988, reaching 20% among Klebsiella pneumoniae and 5% among Escherichia coli isolates by 2004.[11] In response to this, clinicians have increasingly utilized carbapenems to treat patients with serious Gram‐negative infections. However, the development of several types of carbapenemases by Enterobacteriaceae has led to a greater prevalence of carbapenem‐resistant Enterobacteriaceae species (CRE).[12, 13, 14, 15, 16, 17, 18] In fact, a recent report from the Centers for Disease Control and Prevention (CDC) documents a rapid rise in both the prevalence and extent of CRE in the United States.[19]
These Gram‐negative multidrug‐resistant (MDR) organisms frequently cause serious infections including pneumonia and bloodstream infections (BSI). The fact that these conditions, if not addressed in a timely and appropriate manner, lead to high morbidity, mortality, and costs, makes understanding the patterns of resistance that much more critical. To gain a better understanding of the prevalence and characteristics of MDR rates among PA and carbapenem resistance in Enterobacteriaceae in patients hospitalized in the United States with pneumonia and BSI, we conducted a multicenter survey of microbiology data.
METHODS
To determine the prevalence of predefined resistance patterns among PA and Enterobacteriaceae in pneumonia and BSI specimens, we examined The Surveillance Network (TSN) database from Eurofins between years 2000 and 2009. The database has been used extensively for surveillance purposes since 1994 and has previously been described in detail.[17, 20, 21, 22, 23] Briefly, TSN is a warehouse of routine clinical microbiology data collected from a nationally representative sample of microbiology laboratories in 217 hospitals in the United States. To minimize selection bias, laboratories are included based on their geography and the demographics of the populations they serve.[20] Only clinically significant samples are reported. No personal identifying information for source patients is available in this database. Only source laboratories that perform antimicrobial susceptibility testing according standard US Food and Drug Administration‐approved testing methods and interpret susceptibility in accordance with the Clinical Laboratory Standards Institute breakpoints are included.[24] All enrolled laboratories undergo a pre‐enrollment site visit. Logical filters are used for routine quality control to detect unusual susceptibility profiles and to ensure appropriate testing methods. Repeat testing and reporting are done as necessary.[20]
Laboratory samples are reported as susceptible, intermediate, or resistant.[24] We required that samples have susceptibility data for each of the antimicrobials needed to determine their resistance phenotype. These susceptibility patterns served as phenotypic surrogates for resistance. We grouped intermediate samples together with the resistant ones for the purposes of the current analysis. Duplicate isolates were excluded. Only samples representing 1 of the 2 infections of interest, pneumonia and BSI, were included.
We defined MDR‐PA as any isolate resistant to 3 of the following drug classes: aminoglycoside (gentamicin), antipseudomonal penicillin (piperacillin‐tazobactam), antipseudomonal cephalosporin (ceftazidime), carbapenems (imipenem, meropenem), and fluoroquinolone (ciprofloxacin). Enterobacteriaceae were considered CRE if resistant to both a third‐generation cephalosporin and a carbapenem. We examined the data by infection type, year, the 9 US Census geographical divisions, and intensive care unit (ICU) origin.
We did not pursue hypothesis testing due to a high risk of type I error in this large dataset. Therefore, only clinically important trends are highlighted.
RESULTS
Source specimen characteristics for the 205,526 PA (187,343 pneumonia and 18,183 BSI) and 95,566 Enterobacteriaceae specimens (58,810 pneumonia and 36,756 BSI) identified are presented in Table 1. The median age of the patients from which the isolates derive was similar among the PA pneumonia, Enterobacteriaceae pneumonia, and Enterobacteriaceae BSI groups, but higher in the PA BSI group. Similarly, there were differences in the gender distribution of source patients between the organisms and infections. Namely, although females represented a stable 42% of each of the infections with PA, the proportions of females with Enterobacteriaceae pneumonia (36.2%) differed from that in the BSI group (48.6%). Pneumonia specimens (34.0% PA and 39.0% Enterobacteriaceae) were more likely to originate in the ICU than those from BSI (28.4% PA and 21.1% Enterobacteriaceae).
| Pseudomonas aeruginosa, N=205,526 | Enterobacteriaceae, N=95,566 | |
|---|---|---|
| ||
| Pneumonia, n | 187,343 | 58,810 |
| Age, y, median (IQR 25, 75) | 54 (23, 71) | 55 (21, 71) |
| Gender, female, n (%) | 78,418 (41.9) | 21,305 (36.2) |
| ICU origin, n (%) | 63,755 (34.0) | 22,942 (39.0) |
| Meeting definitions of resistance, n (%) | 41,180 (22.0) | 930 (1.6) |
| BSI, n | 18,183 | 36,756 |
| Age, y, median (IQR 25, 75) | 59 (31, 75) | 55 (24, 71) |
| Gender, female, n (%) | 7,448 (41.8) | 17,871 (48.6) |
| ICU origin, n (%) | 5,170 (28.4) | 7,751 (21.1) |
| Meeting definitions of resistance, n (%) | 2,668 (14.7) | 394 (1.1) |
The prevalence of resistance among PA isolates was approximately 15‐fold higher than among Enterobacteriaceae specimens in both infection types (Table 1). This pattern persisted when stratified by infection type (pneumonia: 22.0% MDR‐PA vs 1.6% CRE; BSI: 14.7% MDR‐PA vs 1.1% CRE).
Over the time frame of the study, we detected variable patterns of resistance in the 2 groups of organisms (Figure 1). Namely, among PA in both pneumonia and BSI there was an initial rise in the proportion of MDR specimens between 2000 and 2003, followed by a stabilization until 2005, an additional rise in 2006, and a gradual decline and stabilization through 2009. These fluctuations notwithstanding, there was a net rise in MDR‐PA as a proportion of all PA from 10.7% in 2000 to 13.5% in 2009 among BSI, and from 19.2% in 2000 to 21.7% in 2009 among pneumonia specimens. Among Enterobacteriaceae, the CRE phenotype emerged in 2002 in both infection types and peaked in 2008 at 3.6% in BSI and 5.3% in pneumonia. This peak was followed by a stabilization in 2009 in BSI (3.5%) and a further decline, albeit minor, to 4.6% in pneumonia.

We noted geographic differences in the distribution of resistance (Table 2). Although MDR‐PA was more likely to originate from the East and West North Central divisions, and least likely from the New England and Mountain states, most CRE was detected in the specimens from the latter 2 regions. When stratified by ICU as the location of specimen origin, there were differences in the prevalence of resistant organisms of both types, but these differences were observed only in BSI specimens and not in pneumonia (Figure 2). That is, in BSI, the likelihood of a resistant organism originating from the ICU was approximately double that from a non‐ICU location for both MDR‐PA (21.9% vs 11.8%) and CRE (2.0% vs 0.8%).
| Census Division | MDR‐PA | CRE | ||
|---|---|---|---|---|
| BSI | Pneumonia | BSI | Pneumonia | |
| ||||
| East North Central | 20.8% | 26.9% | 2.0% | 1.9% |
| West North Central | 18.0% | 22.1% | 0.8% | 0.7% |
| East South Central | 15.8% | 20.5% | 0.1% | 0.1% |
| West South Central | 13.5% | 21.7% | 0.3% | 0.5% |
| Pacific | 13.1% | 20.3% | 0.3% | 0.3% |
| Mid‐Atlantic | 12.6% | 20.5% | 2.5% | 3.8% |
| South Atlantic | 12.6% | 21.6% | 0.9% | 1.5% |
| New England | 10.7% | 19.7% | 1.3% | 2.9% |
| Mountain | 8.5% | 19.4% | 0.4% | 1.1% |

DISCUSSION
We have demonstrated that among both pneumonia and BSI specimens, PA and Enterobacteriaceae have a high prevalence of multidrug resistance. When examined cross‐sectionally, in both pneumonia and BSI, the prevalence of MDR‐PA was approximately 15‐fold higher than the prevalence of CRE among Enterobacteriaceae. Over the time frame of the study, MDR‐PA rose and then fell and stabilized to levels only slightly higher than those observed at the beginning of the observation period. In contrast, CRE emerged and rose precipitously between 2006 and 2008, and appeared to stabilize in 2009 in both infection types. Interestingly, we observed geographic variability among resistant isolates. Specifically, the prevalence of CRE was highest in the region with a relatively low prevalence of MDR‐PA. Despite this heterogeneity geographically, resistance for both isolate types in BSI but not in pneumonia was substantially higher in the ICU than outside the ICU.
Our data enhance the current understanding of distribution of Gram‐negative resistance in the United States. A recent study by Braykov and colleagues examined time trends in the development of CRE phenotype among Klebsiella pneumoniae in the United States.[17] By focusing on this single pathogen in various infections within Eurofin's TSN database between 1999 and 2010, they pinpointed its initial emergence to year 2002, with a notably steep rise between 2006 and 2009, with some reduction in the pace of growth in 2010. We have documented an analogous rise in the CRE phenotype among all Enterobacteriaceae, particularly in pneumonia and BSI within a similar time period. Thus, our data on the 1 hand broaden the concern about this pathogen beyond just a single organism within Enterobacteraceae and a single antimicrobial class, and on the other hand serve to focus attention on 2 clinically burdensome infection types, pneumonia and BSI.
Another recent investigation reported a rise in carbapenem‐resistant Enterobacteriaceae in US hospitals over the past decade.[19] Drawing on data from multiple sources, including the dataset used for the current analysis, this study examined the patterns of single‐class resistance to carbapenems among central line‐associated BSI (CLABSI) and catheter‐associated urinary tract infection specimens. Consistent with our findings, these authors noted that the highest percentage of hospitals reporting such single‐class carbapenem‐resistant specimens were located in the Northeastern United States. They also described that the proportion of Enterobacteriaceae with single‐class carbapenem resistance rose from 0% in 2001 to 1.4% in 2010. An additional CDC analysis reported that single‐class carbapenem resistance now exists in 4.2% of Enterobacteraciae as compared to 1.2% of isolates in 2001. We confirm that this rise in single‐class resistance is echoed by a rise in the prevalence of the CRE phenotype, and provides further granularity to this problem, specifically in the setting of pneumonia and BSI.
Although CRE has become an important concern in the treatment of patients with pneumonia and BSI, MDR‐PA remains a far larger challenge in these infections. CREs appear to occur more frequently than in the past but remain relatively dwarfed by the prevalence of MDR‐PA. Our data are generally in agreement with the 2009 to 2010 data from the National Healthcare Safety Network (NHSN) maintained by the CDC, which focuses on CLABSI and ventilator‐associated pneumonia (VAP) rather than general BSI and pneumonia in US hospitals.[25] In this report, the proportion of PA that were classified as MDR according to a definition similar to ours was 15.4% in CLABSI and 17.7% in VAP. In contrast, we document that 13.5% of PA causing BSI and 21.7% causing pneumonia were due to MDR‐PA organisms. This mild divergence likely reflects the slightly different antimicrobials utilized to define MDR‐PA in the 2 studies, as well as variance in the populations examined. An additional data point reported in the NHSN study is the proportion of MDR‐PA CLABSI originating in the ICU (16.8%) versus non‐ICU hospital locations (13.3%). Although the difference we found in the prevalence of BSI by the location in the hospital was greater, we confirm that ICU specimens carry a higher risk of harboring MDR‐PA.
Our study has a number of strengths and limitations. Because we used a nationally representative database to derive our estimates, our results are highly generalizable.
The TSN database consists of microbiology samples from hospital laboratories. Although we attempted to reduce the risk of duplication, because of how samples are numbered in the database, repeat sampling remains a possibility. The definitions of resistance were based on phenotypic patterns of resistance to various antimicrobial classes. This makes our resistant organisms subject to misclassification.
In summary, although carbapenem resistance among Enterobacteriaceae has emerged as an important phenomenon, multidrug resistance among PA remains relatively more prevalent in the United States. Furthermore, over the decade examined, MDR‐PA has remained an important pathogen in pneumonia and BSI that persists across all geographic regions of the United States. Although CRE is rightfully receiving a disproportionate share of attention from public health officials, it would be shortsighted to ignore the importance of MDR‐PA as a target, not only for transmission prevention and antimicrobial stewardship, but also for new therapeutic development. Because the patterns of resistance are rapidly evolving, it is incumbent upon our public health enterprise to perform more granular real‐time surveillance to allow changes in epidemiology to inform policy and treatment decisions.
ACKNOWLEDGEMENTS
Disclosures: This study was supported by a grant from Cubist Pharmaceuticals. The authors report no conflicts of interest.
Administration of initially appropriate antimicrobial therapy represents a key determinant of outcome in patients with severe infection.[1, 2, 3, 4, 5, 6, 7, 8, 9] The variable patterns of antimicrobial resistance seen between and within healthcare institutions complicate the process of antibiotic selection. Although much attention has historically focused on Staphylococcus aureus, resistance among Gram‐negative pathogens has emerged as a major challenge in the care of hospitalized, and particularly critically ill, patients.[2, 10, 11] Multidrug, and more specifically carbapenem resistance, among such common organisms as Pseudomonas aeruginosa (PA) and Enterobacteriaceae represents a major treatment challenge.[2] A recent US‐based surveillance study reported that a quarter of device‐related infections in hospitalized patients were caused by carbapenem‐resistant PA.[10]
In addition to changes in resistance patterns seen among PA isolates, increasing rates of nonsusceptibility have been described among Enterobacteriaceae. Resistance rates to third‐generation cephalosporins in these pathogens have risen steadily since 1988, reaching 20% among Klebsiella pneumoniae and 5% among Escherichia coli isolates by 2004.[11] In response to this, clinicians have increasingly utilized carbapenems to treat patients with serious Gram‐negative infections. However, the development of several types of carbapenemases by Enterobacteriaceae has led to a greater prevalence of carbapenem‐resistant Enterobacteriaceae species (CRE).[12, 13, 14, 15, 16, 17, 18] In fact, a recent report from the Centers for Disease Control and Prevention (CDC) documents a rapid rise in both the prevalence and extent of CRE in the United States.[19]
These Gram‐negative multidrug‐resistant (MDR) organisms frequently cause serious infections including pneumonia and bloodstream infections (BSI). The fact that these conditions, if not addressed in a timely and appropriate manner, lead to high morbidity, mortality, and costs, makes understanding the patterns of resistance that much more critical. To gain a better understanding of the prevalence and characteristics of MDR rates among PA and carbapenem resistance in Enterobacteriaceae in patients hospitalized in the United States with pneumonia and BSI, we conducted a multicenter survey of microbiology data.
METHODS
To determine the prevalence of predefined resistance patterns among PA and Enterobacteriaceae in pneumonia and BSI specimens, we examined The Surveillance Network (TSN) database from Eurofins between years 2000 and 2009. The database has been used extensively for surveillance purposes since 1994 and has previously been described in detail.[17, 20, 21, 22, 23] Briefly, TSN is a warehouse of routine clinical microbiology data collected from a nationally representative sample of microbiology laboratories in 217 hospitals in the United States. To minimize selection bias, laboratories are included based on their geography and the demographics of the populations they serve.[20] Only clinically significant samples are reported. No personal identifying information for source patients is available in this database. Only source laboratories that perform antimicrobial susceptibility testing according standard US Food and Drug Administration‐approved testing methods and interpret susceptibility in accordance with the Clinical Laboratory Standards Institute breakpoints are included.[24] All enrolled laboratories undergo a pre‐enrollment site visit. Logical filters are used for routine quality control to detect unusual susceptibility profiles and to ensure appropriate testing methods. Repeat testing and reporting are done as necessary.[20]
Laboratory samples are reported as susceptible, intermediate, or resistant.[24] We required that samples have susceptibility data for each of the antimicrobials needed to determine their resistance phenotype. These susceptibility patterns served as phenotypic surrogates for resistance. We grouped intermediate samples together with the resistant ones for the purposes of the current analysis. Duplicate isolates were excluded. Only samples representing 1 of the 2 infections of interest, pneumonia and BSI, were included.
We defined MDR‐PA as any isolate resistant to 3 of the following drug classes: aminoglycoside (gentamicin), antipseudomonal penicillin (piperacillin‐tazobactam), antipseudomonal cephalosporin (ceftazidime), carbapenems (imipenem, meropenem), and fluoroquinolone (ciprofloxacin). Enterobacteriaceae were considered CRE if resistant to both a third‐generation cephalosporin and a carbapenem. We examined the data by infection type, year, the 9 US Census geographical divisions, and intensive care unit (ICU) origin.
We did not pursue hypothesis testing due to a high risk of type I error in this large dataset. Therefore, only clinically important trends are highlighted.
RESULTS
Source specimen characteristics for the 205,526 PA (187,343 pneumonia and 18,183 BSI) and 95,566 Enterobacteriaceae specimens (58,810 pneumonia and 36,756 BSI) identified are presented in Table 1. The median age of the patients from which the isolates derive was similar among the PA pneumonia, Enterobacteriaceae pneumonia, and Enterobacteriaceae BSI groups, but higher in the PA BSI group. Similarly, there were differences in the gender distribution of source patients between the organisms and infections. Namely, although females represented a stable 42% of each of the infections with PA, the proportions of females with Enterobacteriaceae pneumonia (36.2%) differed from that in the BSI group (48.6%). Pneumonia specimens (34.0% PA and 39.0% Enterobacteriaceae) were more likely to originate in the ICU than those from BSI (28.4% PA and 21.1% Enterobacteriaceae).
| Pseudomonas aeruginosa, N=205,526 | Enterobacteriaceae, N=95,566 | |
|---|---|---|
| ||
| Pneumonia, n | 187,343 | 58,810 |
| Age, y, median (IQR 25, 75) | 54 (23, 71) | 55 (21, 71) |
| Gender, female, n (%) | 78,418 (41.9) | 21,305 (36.2) |
| ICU origin, n (%) | 63,755 (34.0) | 22,942 (39.0) |
| Meeting definitions of resistance, n (%) | 41,180 (22.0) | 930 (1.6) |
| BSI, n | 18,183 | 36,756 |
| Age, y, median (IQR 25, 75) | 59 (31, 75) | 55 (24, 71) |
| Gender, female, n (%) | 7,448 (41.8) | 17,871 (48.6) |
| ICU origin, n (%) | 5,170 (28.4) | 7,751 (21.1) |
| Meeting definitions of resistance, n (%) | 2,668 (14.7) | 394 (1.1) |
The prevalence of resistance among PA isolates was approximately 15‐fold higher than among Enterobacteriaceae specimens in both infection types (Table 1). This pattern persisted when stratified by infection type (pneumonia: 22.0% MDR‐PA vs 1.6% CRE; BSI: 14.7% MDR‐PA vs 1.1% CRE).
Over the time frame of the study, we detected variable patterns of resistance in the 2 groups of organisms (Figure 1). Namely, among PA in both pneumonia and BSI there was an initial rise in the proportion of MDR specimens between 2000 and 2003, followed by a stabilization until 2005, an additional rise in 2006, and a gradual decline and stabilization through 2009. These fluctuations notwithstanding, there was a net rise in MDR‐PA as a proportion of all PA from 10.7% in 2000 to 13.5% in 2009 among BSI, and from 19.2% in 2000 to 21.7% in 2009 among pneumonia specimens. Among Enterobacteriaceae, the CRE phenotype emerged in 2002 in both infection types and peaked in 2008 at 3.6% in BSI and 5.3% in pneumonia. This peak was followed by a stabilization in 2009 in BSI (3.5%) and a further decline, albeit minor, to 4.6% in pneumonia.

We noted geographic differences in the distribution of resistance (Table 2). Although MDR‐PA was more likely to originate from the East and West North Central divisions, and least likely from the New England and Mountain states, most CRE was detected in the specimens from the latter 2 regions. When stratified by ICU as the location of specimen origin, there were differences in the prevalence of resistant organisms of both types, but these differences were observed only in BSI specimens and not in pneumonia (Figure 2). That is, in BSI, the likelihood of a resistant organism originating from the ICU was approximately double that from a non‐ICU location for both MDR‐PA (21.9% vs 11.8%) and CRE (2.0% vs 0.8%).
| Census Division | MDR‐PA | CRE | ||
|---|---|---|---|---|
| BSI | Pneumonia | BSI | Pneumonia | |
| ||||
| East North Central | 20.8% | 26.9% | 2.0% | 1.9% |
| West North Central | 18.0% | 22.1% | 0.8% | 0.7% |
| East South Central | 15.8% | 20.5% | 0.1% | 0.1% |
| West South Central | 13.5% | 21.7% | 0.3% | 0.5% |
| Pacific | 13.1% | 20.3% | 0.3% | 0.3% |
| Mid‐Atlantic | 12.6% | 20.5% | 2.5% | 3.8% |
| South Atlantic | 12.6% | 21.6% | 0.9% | 1.5% |
| New England | 10.7% | 19.7% | 1.3% | 2.9% |
| Mountain | 8.5% | 19.4% | 0.4% | 1.1% |

DISCUSSION
We have demonstrated that among both pneumonia and BSI specimens, PA and Enterobacteriaceae have a high prevalence of multidrug resistance. When examined cross‐sectionally, in both pneumonia and BSI, the prevalence of MDR‐PA was approximately 15‐fold higher than the prevalence of CRE among Enterobacteriaceae. Over the time frame of the study, MDR‐PA rose and then fell and stabilized to levels only slightly higher than those observed at the beginning of the observation period. In contrast, CRE emerged and rose precipitously between 2006 and 2008, and appeared to stabilize in 2009 in both infection types. Interestingly, we observed geographic variability among resistant isolates. Specifically, the prevalence of CRE was highest in the region with a relatively low prevalence of MDR‐PA. Despite this heterogeneity geographically, resistance for both isolate types in BSI but not in pneumonia was substantially higher in the ICU than outside the ICU.
Our data enhance the current understanding of distribution of Gram‐negative resistance in the United States. A recent study by Braykov and colleagues examined time trends in the development of CRE phenotype among Klebsiella pneumoniae in the United States.[17] By focusing on this single pathogen in various infections within Eurofin's TSN database between 1999 and 2010, they pinpointed its initial emergence to year 2002, with a notably steep rise between 2006 and 2009, with some reduction in the pace of growth in 2010. We have documented an analogous rise in the CRE phenotype among all Enterobacteriaceae, particularly in pneumonia and BSI within a similar time period. Thus, our data on the 1 hand broaden the concern about this pathogen beyond just a single organism within Enterobacteraceae and a single antimicrobial class, and on the other hand serve to focus attention on 2 clinically burdensome infection types, pneumonia and BSI.
Another recent investigation reported a rise in carbapenem‐resistant Enterobacteriaceae in US hospitals over the past decade.[19] Drawing on data from multiple sources, including the dataset used for the current analysis, this study examined the patterns of single‐class resistance to carbapenems among central line‐associated BSI (CLABSI) and catheter‐associated urinary tract infection specimens. Consistent with our findings, these authors noted that the highest percentage of hospitals reporting such single‐class carbapenem‐resistant specimens were located in the Northeastern United States. They also described that the proportion of Enterobacteriaceae with single‐class carbapenem resistance rose from 0% in 2001 to 1.4% in 2010. An additional CDC analysis reported that single‐class carbapenem resistance now exists in 4.2% of Enterobacteraciae as compared to 1.2% of isolates in 2001. We confirm that this rise in single‐class resistance is echoed by a rise in the prevalence of the CRE phenotype, and provides further granularity to this problem, specifically in the setting of pneumonia and BSI.
Although CRE has become an important concern in the treatment of patients with pneumonia and BSI, MDR‐PA remains a far larger challenge in these infections. CREs appear to occur more frequently than in the past but remain relatively dwarfed by the prevalence of MDR‐PA. Our data are generally in agreement with the 2009 to 2010 data from the National Healthcare Safety Network (NHSN) maintained by the CDC, which focuses on CLABSI and ventilator‐associated pneumonia (VAP) rather than general BSI and pneumonia in US hospitals.[25] In this report, the proportion of PA that were classified as MDR according to a definition similar to ours was 15.4% in CLABSI and 17.7% in VAP. In contrast, we document that 13.5% of PA causing BSI and 21.7% causing pneumonia were due to MDR‐PA organisms. This mild divergence likely reflects the slightly different antimicrobials utilized to define MDR‐PA in the 2 studies, as well as variance in the populations examined. An additional data point reported in the NHSN study is the proportion of MDR‐PA CLABSI originating in the ICU (16.8%) versus non‐ICU hospital locations (13.3%). Although the difference we found in the prevalence of BSI by the location in the hospital was greater, we confirm that ICU specimens carry a higher risk of harboring MDR‐PA.
Our study has a number of strengths and limitations. Because we used a nationally representative database to derive our estimates, our results are highly generalizable.
The TSN database consists of microbiology samples from hospital laboratories. Although we attempted to reduce the risk of duplication, because of how samples are numbered in the database, repeat sampling remains a possibility. The definitions of resistance were based on phenotypic patterns of resistance to various antimicrobial classes. This makes our resistant organisms subject to misclassification.
In summary, although carbapenem resistance among Enterobacteriaceae has emerged as an important phenomenon, multidrug resistance among PA remains relatively more prevalent in the United States. Furthermore, over the decade examined, MDR‐PA has remained an important pathogen in pneumonia and BSI that persists across all geographic regions of the United States. Although CRE is rightfully receiving a disproportionate share of attention from public health officials, it would be shortsighted to ignore the importance of MDR‐PA as a target, not only for transmission prevention and antimicrobial stewardship, but also for new therapeutic development. Because the patterns of resistance are rapidly evolving, it is incumbent upon our public health enterprise to perform more granular real‐time surveillance to allow changes in epidemiology to inform policy and treatment decisions.
ACKNOWLEDGEMENTS
Disclosures: This study was supported by a grant from Cubist Pharmaceuticals. The authors report no conflicts of interest.
- National Nosocomial Infections Surveillance (NNIS) System Report. Am J Infect Control. 2004;32:470.
- , , , . National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob Agents Chemother. 2004;48:4606–4610.
- , , , et al. Health care‐associated pneumonia and community‐acquired pneumonia: a single‐center experience. Antimicrob Agents Chemother. 2007;51:3568–3573.
- , , , et al. Clinical importance of delays in the initiation of appropriate antibiotic treatment for ventilator‐associated pneumonia. Chest. 2002;122:262–268.
- . Modification of empiric antibiotic treatment in patients with pneumonia acquired in the intensive care unit. ICU‐Acquired Pneumonia Study Group. Intensive Care Med. 1996;22:387–394.
- , , , , . Antimicrobial therapy escalation and hospital mortality among patients with HCAP: a single center experience. Chest. 2008:134:963–968.
- , , , et al. Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock: 2008. Crit Care Med. 2008;36:296–327.
- , , , , , . Inappropriate antibiotic therapy in Gram‐negative sepsis increases hospital length of stay. Crit Care Med. 2011;39:46–51.
- , , , . Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients. Chest. 1999;115:462–474.
- , , , et al. Antimicrobial‐resistant pathogens associated with healthcare‐associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hospital Epidemiol. 2008;29:996–1011.
- , ; National Nosocomial Infections Surveillance (NNIS) System. Overview of nosocomial infections caused by Gram‐negative bacilli. Clin Infect Dis. 2005;41:848–854.
- , , . The real threat of Klebsiella pneumoniae carbapenemase‐producing bacteria. Lancet Infect Dis. 2009;9:228–236.
- , , , . Household transmission of carbapenemase‐producing Klebsiella pneumoniae. Emerg Infect Dis. 2008;14:859–860.
- , , , . Isolation of imipenem‐resistant Enterobacter species: emergence of KPC‐2 carbapenemase, molecular characterization, epidemiology, and outcomes. Antimicrob Agents Chemother. 2008;52:1413–1418.
- , , , , . Outcomes of carbapenem‐resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008;29:1099–1106.
- , , , , , ; for the Centers for Disease Control and Prevention Epicenter Program. Emergence and rapid regional spread of Klebsiella pneumoniae carbapenemase‐producing Enterobacteriaceae. Clin Infect Dis. 2011;53:532–540.
- , , , , . Trends in resistance to carbapenems and third‐generation cephalosporins among clinical isolates of Klebsiella pneumoniae in the United States, 1999–2010. Infect Control Hosp Epidemiol. 2013;34:259–268.
- , , , . Population‐based incidence of carbapenem‐resistant Klebsiella pneumoniae along the continuum of care, Los Angeles County. Infect Control Hosp Epidemiol. 2013;34:144–150.
- Centers for Disease Control and Prevention (CDC). Vital signs: carbapenem‐resistant enterobacteriaceae. MMWR Morb Mortal Wkly Rep. 2013;62:165–170.
- , , . Antimicrobial resistance in key bloodstream bacterial isolates: electronic surveillance with the Surveillance Network Database–USA. Clin Infect Dis. 1999;29:259–263.
- , , . Community‐associated methicillin‐resistant Staphylococcus aureus in outpatients, United States, 1999–2006. Emerg Infect Dis. 2009;15:1925–1930.
- , , . Increasing resistance of Acinetobacter species to imipenem in United States hospitals, 1999–2006. Infect Control Hosp Epidemiol. 2010;31:196–197.
- , , , , . Prevalence of antimicrobial resistance in bacteria isolated from central nervous system specimens as reported by U.S. hospital laboratories from 2000 to 2002. Ann Clin Microbiol Antimicrob. 2004;3:3.
- Clinical Laboratory Standards Institute. Available at: http://www.clsi.org. Accessed July 8, 2013.
- , , , et al. Antimicrobial‐resistant pathogens associates with healthcare‐associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol. 2013;34:1–14.
- National Nosocomial Infections Surveillance (NNIS) System Report. Am J Infect Control. 2004;32:470.
- , , , . National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob Agents Chemother. 2004;48:4606–4610.
- , , , et al. Health care‐associated pneumonia and community‐acquired pneumonia: a single‐center experience. Antimicrob Agents Chemother. 2007;51:3568–3573.
- , , , et al. Clinical importance of delays in the initiation of appropriate antibiotic treatment for ventilator‐associated pneumonia. Chest. 2002;122:262–268.
- . Modification of empiric antibiotic treatment in patients with pneumonia acquired in the intensive care unit. ICU‐Acquired Pneumonia Study Group. Intensive Care Med. 1996;22:387–394.
- , , , , . Antimicrobial therapy escalation and hospital mortality among patients with HCAP: a single center experience. Chest. 2008:134:963–968.
- , , , et al. Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock: 2008. Crit Care Med. 2008;36:296–327.
- , , , , , . Inappropriate antibiotic therapy in Gram‐negative sepsis increases hospital length of stay. Crit Care Med. 2011;39:46–51.
- , , , . Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients. Chest. 1999;115:462–474.
- , , , et al. Antimicrobial‐resistant pathogens associated with healthcare‐associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hospital Epidemiol. 2008;29:996–1011.
- , ; National Nosocomial Infections Surveillance (NNIS) System. Overview of nosocomial infections caused by Gram‐negative bacilli. Clin Infect Dis. 2005;41:848–854.
- , , . The real threat of Klebsiella pneumoniae carbapenemase‐producing bacteria. Lancet Infect Dis. 2009;9:228–236.
- , , , . Household transmission of carbapenemase‐producing Klebsiella pneumoniae. Emerg Infect Dis. 2008;14:859–860.
- , , , . Isolation of imipenem‐resistant Enterobacter species: emergence of KPC‐2 carbapenemase, molecular characterization, epidemiology, and outcomes. Antimicrob Agents Chemother. 2008;52:1413–1418.
- , , , , . Outcomes of carbapenem‐resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008;29:1099–1106.
- , , , , , ; for the Centers for Disease Control and Prevention Epicenter Program. Emergence and rapid regional spread of Klebsiella pneumoniae carbapenemase‐producing Enterobacteriaceae. Clin Infect Dis. 2011;53:532–540.
- , , , , . Trends in resistance to carbapenems and third‐generation cephalosporins among clinical isolates of Klebsiella pneumoniae in the United States, 1999–2010. Infect Control Hosp Epidemiol. 2013;34:259–268.
- , , , . Population‐based incidence of carbapenem‐resistant Klebsiella pneumoniae along the continuum of care, Los Angeles County. Infect Control Hosp Epidemiol. 2013;34:144–150.
- Centers for Disease Control and Prevention (CDC). Vital signs: carbapenem‐resistant enterobacteriaceae. MMWR Morb Mortal Wkly Rep. 2013;62:165–170.
- , , . Antimicrobial resistance in key bloodstream bacterial isolates: electronic surveillance with the Surveillance Network Database–USA. Clin Infect Dis. 1999;29:259–263.
- , , . Community‐associated methicillin‐resistant Staphylococcus aureus in outpatients, United States, 1999–2006. Emerg Infect Dis. 2009;15:1925–1930.
- , , . Increasing resistance of Acinetobacter species to imipenem in United States hospitals, 1999–2006. Infect Control Hosp Epidemiol. 2010;31:196–197.
- , , , , . Prevalence of antimicrobial resistance in bacteria isolated from central nervous system specimens as reported by U.S. hospital laboratories from 2000 to 2002. Ann Clin Microbiol Antimicrob. 2004;3:3.
- Clinical Laboratory Standards Institute. Available at: http://www.clsi.org. Accessed July 8, 2013.
- , , , et al. Antimicrobial‐resistant pathogens associates with healthcare‐associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol. 2013;34:1–14.
© 2013 Society of Hospital Medicine