User login
Researchers have developed a technique that may help predict whether patients with myelodysplastic syndromes (MDS) or chronic myelomonocytic leukemia (CMML) will respond to treatment with azacytidine (AZA).
“The new method, called AZA-MS, utilizes a cutting-edge technique known as mass spectrometry to measure the different forms of AZA inside blood cells of patients—such as the AZA molecules that are incorporated into the DNA or RNA,” said Ashwin Unnikrishnan, PhD, of the University of New South Wales in Sydney, Australia.
With this method, Dr Unnikrishnan and his colleagues found that patients who do not respond to AZA may incorporate fewer AZA molecules in their DNA and have lower DNA demethylation than responders. However, this is not always the case.
The researchers reported these findings in Leukemia.
The team initially tested AZA-MS in AZA-treated RKO cells and found that AZA-MS could quantify the ribonucleoside (5-AZA-cR) and deoxyribonucleoside (5-AZA-CdR) forms of AZA in RNA, DNA, and the cytoplasm—all in the same sample.
The researchers also found that AZA induced dose-dependent DNA demethylation but did not have an effect on RNA methylation.
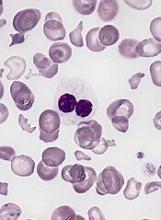
The team then used AZA-MS to analyze bone marrow samples from patients with MDS (n=4) or CMML (n=4) who were undergoing treatment with AZA. All of the patients had received at least 6 cycles of the drug.
Each patient had 3 bone marrow samples collected—one immediately before starting treatment; one on day 8 of cycle 1 (C1d8); and one on day 28 of cycle 1 (C1d28), when they had spent 20 days off the drug.
Four of the patients were complete responders, and 4 were nonresponders. In each group, 2 patients had MDS, and 2 had CMML.
At C1d8, DNA-5-AZA-CdR was significantly greater in responders than nonresponders. And, overall, responders had increased DNA demethylation compared to nonresponders.
However, the researchers also observed differences among the nonresponders. Two nonresponders had very low levels of DNA-5-AZA-CdR at C1d8 and no demethylation. The other 2 nonresponders had much higher DNA-5-AZA-CdR and DNA demethylation levels, which were comparable to levels in responders.
The researchers said they could detect AZA and DNA-5-AZA-CdR intracellularly, as well as RNA-AZA, in the nonresponders with minimal DNA-5-AZA-CdR and DNA demethylation.
The team said this suggests that neither cellular uptake nor intracellular metabolism explain the low DNA-5-AZA-CdR in these patients. Instead, the researchers believe these patients may have a greater proportion of bone marrow cells that are quiescent and not undergoing DNA replication.
The researchers also believe the nonresponders with higher DNA-5-AZA-CdR may be explained by a failure to induce an interferon response, which is necessary for a clinical response.
On the other hand, these nonresponders could have defective immune cell-mediated clearance of dysplastic cells or increased tolerance to this clearance, the researchers said.
The team also noted that, at C1d28, DNA-5-AZA-CdR levels dropped (but were still detectable) in all 8 patients, and DNA methylation had nearly returned to pretreatment levels in all patients. ![]()
Researchers have developed a technique that may help predict whether patients with myelodysplastic syndromes (MDS) or chronic myelomonocytic leukemia (CMML) will respond to treatment with azacytidine (AZA).
“The new method, called AZA-MS, utilizes a cutting-edge technique known as mass spectrometry to measure the different forms of AZA inside blood cells of patients—such as the AZA molecules that are incorporated into the DNA or RNA,” said Ashwin Unnikrishnan, PhD, of the University of New South Wales in Sydney, Australia.
With this method, Dr Unnikrishnan and his colleagues found that patients who do not respond to AZA may incorporate fewer AZA molecules in their DNA and have lower DNA demethylation than responders. However, this is not always the case.
The researchers reported these findings in Leukemia.
The team initially tested AZA-MS in AZA-treated RKO cells and found that AZA-MS could quantify the ribonucleoside (5-AZA-cR) and deoxyribonucleoside (5-AZA-CdR) forms of AZA in RNA, DNA, and the cytoplasm—all in the same sample.
The researchers also found that AZA induced dose-dependent DNA demethylation but did not have an effect on RNA methylation.
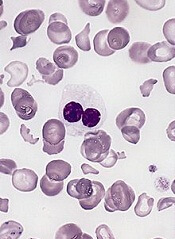
The team then used AZA-MS to analyze bone marrow samples from patients with MDS (n=4) or CMML (n=4) who were undergoing treatment with AZA. All of the patients had received at least 6 cycles of the drug.
Each patient had 3 bone marrow samples collected—one immediately before starting treatment; one on day 8 of cycle 1 (C1d8); and one on day 28 of cycle 1 (C1d28), when they had spent 20 days off the drug.
Four of the patients were complete responders, and 4 were nonresponders. In each group, 2 patients had MDS, and 2 had CMML.
At C1d8, DNA-5-AZA-CdR was significantly greater in responders than nonresponders. And, overall, responders had increased DNA demethylation compared to nonresponders.
However, the researchers also observed differences among the nonresponders. Two nonresponders had very low levels of DNA-5-AZA-CdR at C1d8 and no demethylation. The other 2 nonresponders had much higher DNA-5-AZA-CdR and DNA demethylation levels, which were comparable to levels in responders.
The researchers said they could detect AZA and DNA-5-AZA-CdR intracellularly, as well as RNA-AZA, in the nonresponders with minimal DNA-5-AZA-CdR and DNA demethylation.
The team said this suggests that neither cellular uptake nor intracellular metabolism explain the low DNA-5-AZA-CdR in these patients. Instead, the researchers believe these patients may have a greater proportion of bone marrow cells that are quiescent and not undergoing DNA replication.
The researchers also believe the nonresponders with higher DNA-5-AZA-CdR may be explained by a failure to induce an interferon response, which is necessary for a clinical response.
On the other hand, these nonresponders could have defective immune cell-mediated clearance of dysplastic cells or increased tolerance to this clearance, the researchers said.
The team also noted that, at C1d28, DNA-5-AZA-CdR levels dropped (but were still detectable) in all 8 patients, and DNA methylation had nearly returned to pretreatment levels in all patients. ![]()
Researchers have developed a technique that may help predict whether patients with myelodysplastic syndromes (MDS) or chronic myelomonocytic leukemia (CMML) will respond to treatment with azacytidine (AZA).
“The new method, called AZA-MS, utilizes a cutting-edge technique known as mass spectrometry to measure the different forms of AZA inside blood cells of patients—such as the AZA molecules that are incorporated into the DNA or RNA,” said Ashwin Unnikrishnan, PhD, of the University of New South Wales in Sydney, Australia.
With this method, Dr Unnikrishnan and his colleagues found that patients who do not respond to AZA may incorporate fewer AZA molecules in their DNA and have lower DNA demethylation than responders. However, this is not always the case.
The researchers reported these findings in Leukemia.
The team initially tested AZA-MS in AZA-treated RKO cells and found that AZA-MS could quantify the ribonucleoside (5-AZA-cR) and deoxyribonucleoside (5-AZA-CdR) forms of AZA in RNA, DNA, and the cytoplasm—all in the same sample.
The researchers also found that AZA induced dose-dependent DNA demethylation but did not have an effect on RNA methylation.
The team then used AZA-MS to analyze bone marrow samples from patients with MDS (n=4) or CMML (n=4) who were undergoing treatment with AZA. All of the patients had received at least 6 cycles of the drug.
Each patient had 3 bone marrow samples collected—one immediately before starting treatment; one on day 8 of cycle 1 (C1d8); and one on day 28 of cycle 1 (C1d28), when they had spent 20 days off the drug.
Four of the patients were complete responders, and 4 were nonresponders. In each group, 2 patients had MDS, and 2 had CMML.
At C1d8, DNA-5-AZA-CdR was significantly greater in responders than nonresponders. And, overall, responders had increased DNA demethylation compared to nonresponders.
However, the researchers also observed differences among the nonresponders. Two nonresponders had very low levels of DNA-5-AZA-CdR at C1d8 and no demethylation. The other 2 nonresponders had much higher DNA-5-AZA-CdR and DNA demethylation levels, which were comparable to levels in responders.
The researchers said they could detect AZA and DNA-5-AZA-CdR intracellularly, as well as RNA-AZA, in the nonresponders with minimal DNA-5-AZA-CdR and DNA demethylation.
The team said this suggests that neither cellular uptake nor intracellular metabolism explain the low DNA-5-AZA-CdR in these patients. Instead, the researchers believe these patients may have a greater proportion of bone marrow cells that are quiescent and not undergoing DNA replication.
The researchers also believe the nonresponders with higher DNA-5-AZA-CdR may be explained by a failure to induce an interferon response, which is necessary for a clinical response.
On the other hand, these nonresponders could have defective immune cell-mediated clearance of dysplastic cells or increased tolerance to this clearance, the researchers said.
The team also noted that, at C1d28, DNA-5-AZA-CdR levels dropped (but were still detectable) in all 8 patients, and DNA methylation had nearly returned to pretreatment levels in all patients. ![]()