User login
As the nation’s largest integrated health care system with about 50,000 new cancer diagnoses per year, providing care for over 400,000 veterans with cancer and a robust research portfolio, the US Department of Veterans Affairs (VA) is well positioned to be a leader in both clinical and research in oncology. The VA National Precision Oncology Program (NPOP), which provides tumor sequencing and consultative services, is a key component of VA oncology assets.
Case Presentation
As the mission of the VA is to “care for him who shall have borne the battle,” it is fitting to begin with the story of a US Army veteran in his 40s and the father of 2 young children who developed progressive shortness of breath, cough, and weight loss over a period of 8 months. He was diagnosed with metastatic lung adenocarcinoma in 2016, and standard testing of his tumor showed no alteration of the EGFR and ALK genes. He was treated with whole brain radiation and had begun treatment for carboplatin and pemetrexed chemotherapy with mixed tumor response.
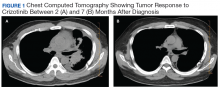
Subsequently, his tumor was tested through NPOP, using a multigene next-generation sequencing (NGS) assay panel, which showed the presence of an abnormal fusion between the EML4 and ALK genes. The chemotherapy was discontinued and oral crizotinib precision therapy was started. The patient had an excellent response in all sites of disease (Figure 1). He was able to return to work and school.
In July 2017, his medication was switched to alectinib for asymptomatic progression in his brain, and there was further response. In September 2019, he was treated with precision intensity-modulated radiotherapy (IMRT), targeting a single brain metastasis as there were no other sites of cancer progression and no cancerrelated symptoms. He finished school and continues to work.
Precision Oncology
Oncology is a relatively young medical field. The early medical treatments for cancer were developed empirically against hematologic malignancies, particularly leukemias. Cytotoxic chemotherapeutic agents as a group have modest effects on most solid tumors, and even modern genomics has had limited ability to predict differential benefit in patients with advanced-stage carcinomas. As a result, the medications are used in a nonprecision manner in which all patients with the same cancer diagnosis and stage receive the same treatment. This is due in part to our limited understanding of both the pathophysiology of cancer and the mechanism of action of cytotoxic agents.
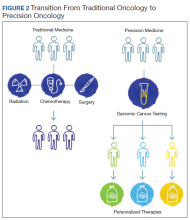
The paradigm of precision oncology, in contrast, utilizes unique, patient-specific molecular characteristics to guide prescribing of antineoplastic agents (Figure 2). These molecular characteristics are frequently tumoral but also may be nontumoral, such as germline genetic variants and even nonhuman, such as the gut microbiome as has been proposed as predictive of response to immune checkpoint inhibitors.1,2
One of the first examples of precision oncology was tumor testing for the estrogen receptor in breast cancer, which distinguishes breast tumors sensitive to hormonal treatments from those that are resistant.3 In 2004, somatically acquired mutation of the EGFR gene was found to be associated with response to EGFR tyrosine kinase inhibitors such as gefitinib and erlotinib, and subsequently it was shown that patients without these mutations derived no benefit from use of these drugs.4 Thus, the precision oncology paradigm is using a molecular diagnostic as part of the indication for an antineoplastic agent, resulting in improved therapeutic efficacy and often reduced toxicity.
By 2015, multiple examples of DNA-based gene alterations that predict drug response were known, including at least 5 in non-small cell lung cancer (NSCLC). The heterogeneity of molecular testing practice patterns and methods of testing in VA along with the increasing number and complexity of molecular tests facilitated launch of a regional precision oncology program based primarily in Veterans Integrated Service Network 1, which provided tumor DNA sequencing through 2 vendors. Advances in DNA sequencing technology, particularly NGS, permit sequencing of multiple genes in clinical tumor samples, using a panel applicable for multiple tumor types. As part of VA contributions to the 2016 White House Cancer Moonshot initiative, the regional program became NPOP with expanded geographic scope, the addition of clinical consultative services, and robust informatics that supports associated research and a learning health care system. NPOP is a component of the VA National Oncology Program Office under the Office of Specialty Care.
Testing
With the launch of NPOP in mid-2016, there was rapid expansion of the number of VA facilities participating, and the number of tumor samples being submitted increased substantially. 5 The expansion was facilitated by both central funding for the tumor DNA sequencing and by NPOP-provided training of pathology laboratory staff and oncologists. Today, NPOP is utilized by almost every oncology practice in VA.
NPOP’s initial focus was on lung cancer, specifically advanced-stage nonsquamous NSCLC, which not only is very common in VA, but also has one of the highest number of mutated genes that result in sensitivity to antineoplastic drugs. Recently, metastatic prostate cancer was added as a second focus tumor type. Dashboards are available on the NPOP website to assist care teams in identifying veterans at their facility with either lung or prostate cancer who may be appropriate for testing. Other solid tumors can be sent for testing through NPOP if patients have advanced stage cancer and are medically appropriate for antineoplastic therapy. To date, NPOP has sequenced > 13,000 samples.
Testing options have been added to NPOP in addition to tumor DNA sequencing. The first addition was the so-called liquid biopsy, more properly known as the cell-free DNA (cfDNA) test, a plasma-based high-sensitivity DNA sequencing assay. cfDNA is shed from dying cells and can be captured and sequenced from a plasma sample obtained by standard venipuncture, using a special-purpose sample collection tube. The test is appropriate for patients who do not have an appropriate archival tumor sample or those who cannot have a new biopsy of tumor tissue. Tumor tissue remains the preferred test sample due to a higher sensitivity than that of cfDNA and less susceptibility to false positives, so consideration of a tumor biopsy is appropriate prior to requesting a cfDNA assay. Therapy can greatly impact the sensitivity of cfDNA testing, so patients should be having disease progression at the time of obtaining a blood sample for cfDNA.
Finally, myeloid leukocytic cells accumulate genetic alterations during aging similar to those found in myelodysplasia and acute myeloid leukemia. These myeloid-associated mutations can be detected in both tumor and cfDNA samples and are known as clonal hyperplasia of indeterminate potential (CHIP). CHIP is much more common in the cfDNA. For lung cancer, CHIP-associated gene variants are readily distinguished from lung cancer-associated variants, but that distinction is much more difficult in many other tumor types.
In partnership with the current DNA sequencing contractor, NPOP provides access to a second gene panel for hematologic malignancies or sarcomas, though neither of these classes of malignancies currently have clear indications for routine NGS multigene panel testing. Given the low rate of finding a gene mutation that would change therapy that could not be found with smaller, less expensive gene panels, NPOP requires prior approval for the use of this panel.
Finally, since early 2019, programmed deathligand 1 (PD-L1) immunohistochemistry analysis is available through NPOP in association with NGS testing of the same sample for those solid tumors with US Food and Drug Administration (FDA)-approved indications that include a PD-L1 companion diagnostic. This service was added to facilitate concurrent testing of PD-L1 and DNA sequencing, which speeds availability of molecular data to the health care provider and veteran.
Determining Clinical Significance
The complexity of tumor NGS gene panel test results is far greater than frequently ordered laboratory or molecular testing due to the near infinite number of possible results and varying degrees of consensus of the significance of the results for therapeutic decision making. That complexity is reflected in the length of the test reports, which are often ≥ 20 pages. Starting from the gene variants identified by the DNA sequencing variant-caller bioinformatics pipeline, there is a 2-step process, referred to as annotation, to interpret the clinical significance that is repeated for each variant.
The first step is to assign a pathogenicity value, also known as oncogenicity, using a 5-point Likert scale from pathogenic to benign with variant of unknown significance (VUS) in the middle of the scale. Only variants that are pathogenic or likely pathogenic are considered further. A VUS is usually communicated to the health care provider but should generally not be acted on, while benign and likely benign variants may or may not be included in the report and should never be acted on. NPOP examined the concordance of pathogenicity calls among 3 annotation services: N-of-One/QCI Precision Insights (qiagen.com), IBM Watson for Genomics (WfG), and OncoKB (www.oncokb.org).6 There was moderate-to-poor concordance, indicating lack of consensus about whether a significant fraction of observed gene variants contributes to the patient’s cancer. This variability likely arises due to differences in algorithms and criteria used to assess pathogenicity.
The second step of annotation is assignment of the actionability of the variant, using a level of evidence (LoE) scale from 1 (on-label indication) to 4 (absence of clinical evidence; ie, only preclinical or theoretical evidence). Initially, NPOP used an adaptation of the LoE scales from WfG and OncoKB but now mostly uses the recently revised OncoKB LoE. Actionability also includes prediction of resistance to a treatment (LoE level R1 and R2). An example of a resistance gene variant is a KRAS mutation in colorectal cancer, which predicts lack of clinical benefit from anti- EGFR antibodies. It is important to note that a determination of actionability requires 3 inputs: gene, variant, and tumor type. A BRAF V600E mutation in melanoma has different medications with level 1 LoE than does the same mutation in colorectal cancer, for example.
Another complexity in annotation for actionability is tumor type ontogeny—the classification system used for cancer types. WfG uses a subset of the National Cancer Institute Thesaurus (ncithesaurus.nci.nih.gov), OncoKB uses the unique OncoTree (oncotree.mskcc.org), and Foundation Medicine (www.foundationmed icine.com), and N-of-One use propriety classification systems. The WfG and OncoKB tumor types have evolved over time, while it is unclear what changes have been made in the FMI and N-of-One tumor type classification systems. Thus, a gene variant observed in a single patient may be annotated differently by these services because of how the tumor type is mapped onto the services’ tumor type ontogeny. NPOP has been assigning WfG diagnoses since 2017, including historic assignment for prior samples back to the pilot project in 2015. In early 2019, NPOP began requiring test requesters to include International Classification of Diseases for Oncology, 3rd Edition (ICD-O-3) diagnoses (histology and site codes) with the sample. ICD-O-3 codes are used in all cancer registry data in North America, including the VA Cancer Registry System. The approximately 50,000 possible diagnoses allow fine precision in diagnoses, which is important for less common and rare cancer types; however, the large number of diagnoses adds complexity. NPOP has created a partial translation table for ICD-O-3 to WfG diagnosis that includes all diagnoses seen to date; this table facilitates continuing provision of WfG diagnosis without manual review as was previously required.
NPOP-Provided Genetic Services
Given these complexities in interpretation of NGS multigene panel results, NPOP provides several services to assist health care providers and other members of the care team. First, the NPOP Interfacility Consult (IFC) is a virtual “phone-a-friend” service that provides asynchronous patient-specific expert recommendations in precision oncology. By far the most requested service is assistance with interpretation of a patient’s DNA sequence results. Other requests include advice on whether to perform NGS testing and what molecular testing to perform. The IFC is integral to the VA Computerized Patient Record System electronic health record. Additional requests have been submitted and answered by e-mail.
The Molecular Oncology Tumor Board is a monthly case-based educational conference supported by the VA Employee Education Service, which provides continuing education credits for attendees. NPOP staff coordinate the conference, and a panel of specialists from around the country provide expert commentary.
In 2016, IBM gifted the services of WfG to VA. WfG’s main functionality is annotation of NGS results. About 5,000 samples were processed from 2017 to 2019; sample processing is expected to resume shortly. The availability of WfG annotations early in NPOP operation was very useful to the implementation of NPOP in general and the NPOP consultation services in particular, resulting in improved thoroughness of opinions provided by NPOP staff.
Informatics
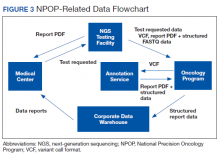
Informatics is an essential component of NPOP that facilitates both clinical care and research (Figure 3). Results of NGS gene panels are returned to the facility that submitted the sample for testing as a PDF document. NPOP receives the same PDF report in real time but also structured data of the results including a variant callformat file and XML file. The secondary sequence data in binary alignment map or FASTQ format is received in batches. NPOP program staff extract data from these files and then load it into SQL tables in the VA Corporate Data Warehouse. In partnership with the VA Pharmacy Benefits Management Service, NPOP has constructed user-friendly dashboards that allow users with no technical skills and who have the appropriate data access permissions to view various portions of the NPOP database. There are dashboards to display a list of NPOP samples by facility, find a patient by name or other identifying information, and display a list of patients who have received any antineoplastic drug, among other functions.
The NPOP database has been used to reannotate NGS results, such as when new drugs are approved. For example, when the first neurotrophic tropomyosin receptor kinase (NTRK) inhibitor was approved, NPOP rapidly identified all living patients with NTRK fusions and notified the health care providers of the availability a potential new treatment option for their patient. 7 NPOP is now building a method to allow NPOP dashboard users to similarly identify patients who have not received a corresponding drug for a user-selected LoE annotation. This will facilitate a systems approach to ensure that all patients with EGFR exon 19 deletions, for example, either have received an EGFR inhibitor or are reviewed to determine why they have not. Furthermore, the database will facilitate real-world data analysis in precision oncology. For example, prior to the recent FDA-approval of poly–(adenosine diphosphate–ribose) polymerase (PARP) inhibitors for prostate cancer, 50 veterans already had been treated with one of these agents. These data can help further inform which of the many variants of DNA damage repair genes benefit from PARP inhibitors.
Ensuring Access to Care for All Veterans
With any new medical technology comes an obligation to ensure appropriate equal access so as to not exacerbate health care disparities. Veterans enrolled in VA health care are much more likely to live in rural communities than does the US population as a whole, and there was concern that these veterans would not receive NGS testing at the same rate as urban veterans. NPOP therefore was intentional during implementation to ensure rural veterans were being offered testing. Indeed, there has been nearly equal utilization by rurality. No other disparities in NPOP utilization have been seen.
A majority of veterans who undergo testing in NPOP have at least 1 actionable gene variant reported.5 However, some of these are for lower LoE off-label use of FDA-approved medications, but many are for agents available only through clinical trials. Consideration of treatments available through a clinical trial is part of standard practice for patients with advanced malignancies. NPOP data have helped identify cohorts who are eligible for clinical trials on the basis of their tumor DNA sequencing results. The National Oncology Program Office has been working closely with the VA Office of Research and Development to expand access to cancer clinical trials in VA. Veterans also can be treated on trials outside VA as medically appropriate and with local VA approval.
Conclusions
The VA NPOP is one of the largest clinical DNA sequencing programs in the nation with integrated consultation services and health informatics resources to facilitate patient care, clinical trials, and health outcomes research. The clinical services of NPOP provide cuttingedge oncology services to veterans throughout VA without exacerbating disparities and will be a national resource for research.
Acknowledgments
NPOP was made possible and implemented through the efforts of a number of people in VHA, including the national and regional leaders who supported the program’s vision and implementation, especially Michael Mayo-Smith, David Shulkin, Jennifer S. Lee, and Laurence Meyer, the leaders and staff of the Massachusetts Veterans Epidemiology Research and Information Center who piloted regional NGS testing, and especially my current and former colleagues in the VA National Oncology Program Office, without whom NPOP would not be possible. The contributions of Neil L. Spector who served as inaugural Director of Precision Oncology and Jill E. Duffy in her role as Director of Oncology Operations are particularly noteworthy.
1. Lima ZS, Ghadamzadeh M, Arashloo FT, Amjad G, Ebadi MR, Younesi L. Recent advances of therapeutic targets based on the molecular signature in breast cancer: genetic mutations and implications for current treatment paradigms. J Hematol Oncol. 2019;12(1):38. Published 2019 Apr 11. doi:10.1186/s13045-019-0725-6
2. Fessler J, Matson V, Gajewski TF. Exploring the emerging role of the microbiome in cancer immunotherapy. J Immunother Cancer. 2019;7(1):108. Published 2019 Apr 17. doi:10.1186/s40425-019-0574-4
3. Kiang DT, Kennedy BJ. Tamoxifen (antiestrogen) therapy in advanced breast cancer. Ann Intern Med. 1977;87(6):687- 690. doi:10.7326/0003-4819-87-6-687.
4. Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497-1500. doi:10.1126/science.1099314
5. Poonnen P, Duffy J, Hintze BJ, et al. Genomic analysis of metastatic solid tumors in veterans: findings from the VHA National Precision Oncology Program. J Clin Oncol. 2019;37(suppl 15):3074. doi:10.1200/JCO.2019.37.15_suppl.3074
6. Katsoulakis E, Duffy JE, Hintze B, Spector NL, Kelley MJ. Comparison of annotation services for nextgeneration sequencing in a large-scale precision oncology program. JCO Precis Oncol. 2020(4):212-221. doi:10.1200/PO.19.00118
7. Drilon A, Laetsch TW, Kummar S, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378(8):731-739. doi:10.1056/NEJMoa1714448
As the nation’s largest integrated health care system with about 50,000 new cancer diagnoses per year, providing care for over 400,000 veterans with cancer and a robust research portfolio, the US Department of Veterans Affairs (VA) is well positioned to be a leader in both clinical and research in oncology. The VA National Precision Oncology Program (NPOP), which provides tumor sequencing and consultative services, is a key component of VA oncology assets.
Case Presentation
As the mission of the VA is to “care for him who shall have borne the battle,” it is fitting to begin with the story of a US Army veteran in his 40s and the father of 2 young children who developed progressive shortness of breath, cough, and weight loss over a period of 8 months. He was diagnosed with metastatic lung adenocarcinoma in 2016, and standard testing of his tumor showed no alteration of the EGFR and ALK genes. He was treated with whole brain radiation and had begun treatment for carboplatin and pemetrexed chemotherapy with mixed tumor response.
Subsequently, his tumor was tested through NPOP, using a multigene next-generation sequencing (NGS) assay panel, which showed the presence of an abnormal fusion between the EML4 and ALK genes. The chemotherapy was discontinued and oral crizotinib precision therapy was started. The patient had an excellent response in all sites of disease (Figure 1). He was able to return to work and school.
In July 2017, his medication was switched to alectinib for asymptomatic progression in his brain, and there was further response. In September 2019, he was treated with precision intensity-modulated radiotherapy (IMRT), targeting a single brain metastasis as there were no other sites of cancer progression and no cancerrelated symptoms. He finished school and continues to work.
Precision Oncology
Oncology is a relatively young medical field. The early medical treatments for cancer were developed empirically against hematologic malignancies, particularly leukemias. Cytotoxic chemotherapeutic agents as a group have modest effects on most solid tumors, and even modern genomics has had limited ability to predict differential benefit in patients with advanced-stage carcinomas. As a result, the medications are used in a nonprecision manner in which all patients with the same cancer diagnosis and stage receive the same treatment. This is due in part to our limited understanding of both the pathophysiology of cancer and the mechanism of action of cytotoxic agents.
The paradigm of precision oncology, in contrast, utilizes unique, patient-specific molecular characteristics to guide prescribing of antineoplastic agents (Figure 2). These molecular characteristics are frequently tumoral but also may be nontumoral, such as germline genetic variants and even nonhuman, such as the gut microbiome as has been proposed as predictive of response to immune checkpoint inhibitors.1,2
One of the first examples of precision oncology was tumor testing for the estrogen receptor in breast cancer, which distinguishes breast tumors sensitive to hormonal treatments from those that are resistant.3 In 2004, somatically acquired mutation of the EGFR gene was found to be associated with response to EGFR tyrosine kinase inhibitors such as gefitinib and erlotinib, and subsequently it was shown that patients without these mutations derived no benefit from use of these drugs.4 Thus, the precision oncology paradigm is using a molecular diagnostic as part of the indication for an antineoplastic agent, resulting in improved therapeutic efficacy and often reduced toxicity.
By 2015, multiple examples of DNA-based gene alterations that predict drug response were known, including at least 5 in non-small cell lung cancer (NSCLC). The heterogeneity of molecular testing practice patterns and methods of testing in VA along with the increasing number and complexity of molecular tests facilitated launch of a regional precision oncology program based primarily in Veterans Integrated Service Network 1, which provided tumor DNA sequencing through 2 vendors. Advances in DNA sequencing technology, particularly NGS, permit sequencing of multiple genes in clinical tumor samples, using a panel applicable for multiple tumor types. As part of VA contributions to the 2016 White House Cancer Moonshot initiative, the regional program became NPOP with expanded geographic scope, the addition of clinical consultative services, and robust informatics that supports associated research and a learning health care system. NPOP is a component of the VA National Oncology Program Office under the Office of Specialty Care.
Testing
With the launch of NPOP in mid-2016, there was rapid expansion of the number of VA facilities participating, and the number of tumor samples being submitted increased substantially. 5 The expansion was facilitated by both central funding for the tumor DNA sequencing and by NPOP-provided training of pathology laboratory staff and oncologists. Today, NPOP is utilized by almost every oncology practice in VA.
NPOP’s initial focus was on lung cancer, specifically advanced-stage nonsquamous NSCLC, which not only is very common in VA, but also has one of the highest number of mutated genes that result in sensitivity to antineoplastic drugs. Recently, metastatic prostate cancer was added as a second focus tumor type. Dashboards are available on the NPOP website to assist care teams in identifying veterans at their facility with either lung or prostate cancer who may be appropriate for testing. Other solid tumors can be sent for testing through NPOP if patients have advanced stage cancer and are medically appropriate for antineoplastic therapy. To date, NPOP has sequenced > 13,000 samples.
Testing options have been added to NPOP in addition to tumor DNA sequencing. The first addition was the so-called liquid biopsy, more properly known as the cell-free DNA (cfDNA) test, a plasma-based high-sensitivity DNA sequencing assay. cfDNA is shed from dying cells and can be captured and sequenced from a plasma sample obtained by standard venipuncture, using a special-purpose sample collection tube. The test is appropriate for patients who do not have an appropriate archival tumor sample or those who cannot have a new biopsy of tumor tissue. Tumor tissue remains the preferred test sample due to a higher sensitivity than that of cfDNA and less susceptibility to false positives, so consideration of a tumor biopsy is appropriate prior to requesting a cfDNA assay. Therapy can greatly impact the sensitivity of cfDNA testing, so patients should be having disease progression at the time of obtaining a blood sample for cfDNA.
Finally, myeloid leukocytic cells accumulate genetic alterations during aging similar to those found in myelodysplasia and acute myeloid leukemia. These myeloid-associated mutations can be detected in both tumor and cfDNA samples and are known as clonal hyperplasia of indeterminate potential (CHIP). CHIP is much more common in the cfDNA. For lung cancer, CHIP-associated gene variants are readily distinguished from lung cancer-associated variants, but that distinction is much more difficult in many other tumor types.
In partnership with the current DNA sequencing contractor, NPOP provides access to a second gene panel for hematologic malignancies or sarcomas, though neither of these classes of malignancies currently have clear indications for routine NGS multigene panel testing. Given the low rate of finding a gene mutation that would change therapy that could not be found with smaller, less expensive gene panels, NPOP requires prior approval for the use of this panel.
Finally, since early 2019, programmed deathligand 1 (PD-L1) immunohistochemistry analysis is available through NPOP in association with NGS testing of the same sample for those solid tumors with US Food and Drug Administration (FDA)-approved indications that include a PD-L1 companion diagnostic. This service was added to facilitate concurrent testing of PD-L1 and DNA sequencing, which speeds availability of molecular data to the health care provider and veteran.
Determining Clinical Significance
The complexity of tumor NGS gene panel test results is far greater than frequently ordered laboratory or molecular testing due to the near infinite number of possible results and varying degrees of consensus of the significance of the results for therapeutic decision making. That complexity is reflected in the length of the test reports, which are often ≥ 20 pages. Starting from the gene variants identified by the DNA sequencing variant-caller bioinformatics pipeline, there is a 2-step process, referred to as annotation, to interpret the clinical significance that is repeated for each variant.
The first step is to assign a pathogenicity value, also known as oncogenicity, using a 5-point Likert scale from pathogenic to benign with variant of unknown significance (VUS) in the middle of the scale. Only variants that are pathogenic or likely pathogenic are considered further. A VUS is usually communicated to the health care provider but should generally not be acted on, while benign and likely benign variants may or may not be included in the report and should never be acted on. NPOP examined the concordance of pathogenicity calls among 3 annotation services: N-of-One/QCI Precision Insights (qiagen.com), IBM Watson for Genomics (WfG), and OncoKB (www.oncokb.org).6 There was moderate-to-poor concordance, indicating lack of consensus about whether a significant fraction of observed gene variants contributes to the patient’s cancer. This variability likely arises due to differences in algorithms and criteria used to assess pathogenicity.
The second step of annotation is assignment of the actionability of the variant, using a level of evidence (LoE) scale from 1 (on-label indication) to 4 (absence of clinical evidence; ie, only preclinical or theoretical evidence). Initially, NPOP used an adaptation of the LoE scales from WfG and OncoKB but now mostly uses the recently revised OncoKB LoE. Actionability also includes prediction of resistance to a treatment (LoE level R1 and R2). An example of a resistance gene variant is a KRAS mutation in colorectal cancer, which predicts lack of clinical benefit from anti- EGFR antibodies. It is important to note that a determination of actionability requires 3 inputs: gene, variant, and tumor type. A BRAF V600E mutation in melanoma has different medications with level 1 LoE than does the same mutation in colorectal cancer, for example.
Another complexity in annotation for actionability is tumor type ontogeny—the classification system used for cancer types. WfG uses a subset of the National Cancer Institute Thesaurus (ncithesaurus.nci.nih.gov), OncoKB uses the unique OncoTree (oncotree.mskcc.org), and Foundation Medicine (www.foundationmed icine.com), and N-of-One use propriety classification systems. The WfG and OncoKB tumor types have evolved over time, while it is unclear what changes have been made in the FMI and N-of-One tumor type classification systems. Thus, a gene variant observed in a single patient may be annotated differently by these services because of how the tumor type is mapped onto the services’ tumor type ontogeny. NPOP has been assigning WfG diagnoses since 2017, including historic assignment for prior samples back to the pilot project in 2015. In early 2019, NPOP began requiring test requesters to include International Classification of Diseases for Oncology, 3rd Edition (ICD-O-3) diagnoses (histology and site codes) with the sample. ICD-O-3 codes are used in all cancer registry data in North America, including the VA Cancer Registry System. The approximately 50,000 possible diagnoses allow fine precision in diagnoses, which is important for less common and rare cancer types; however, the large number of diagnoses adds complexity. NPOP has created a partial translation table for ICD-O-3 to WfG diagnosis that includes all diagnoses seen to date; this table facilitates continuing provision of WfG diagnosis without manual review as was previously required.
NPOP-Provided Genetic Services
Given these complexities in interpretation of NGS multigene panel results, NPOP provides several services to assist health care providers and other members of the care team. First, the NPOP Interfacility Consult (IFC) is a virtual “phone-a-friend” service that provides asynchronous patient-specific expert recommendations in precision oncology. By far the most requested service is assistance with interpretation of a patient’s DNA sequence results. Other requests include advice on whether to perform NGS testing and what molecular testing to perform. The IFC is integral to the VA Computerized Patient Record System electronic health record. Additional requests have been submitted and answered by e-mail.
The Molecular Oncology Tumor Board is a monthly case-based educational conference supported by the VA Employee Education Service, which provides continuing education credits for attendees. NPOP staff coordinate the conference, and a panel of specialists from around the country provide expert commentary.
In 2016, IBM gifted the services of WfG to VA. WfG’s main functionality is annotation of NGS results. About 5,000 samples were processed from 2017 to 2019; sample processing is expected to resume shortly. The availability of WfG annotations early in NPOP operation was very useful to the implementation of NPOP in general and the NPOP consultation services in particular, resulting in improved thoroughness of opinions provided by NPOP staff.
Informatics
Informatics is an essential component of NPOP that facilitates both clinical care and research (Figure 3). Results of NGS gene panels are returned to the facility that submitted the sample for testing as a PDF document. NPOP receives the same PDF report in real time but also structured data of the results including a variant callformat file and XML file. The secondary sequence data in binary alignment map or FASTQ format is received in batches. NPOP program staff extract data from these files and then load it into SQL tables in the VA Corporate Data Warehouse. In partnership with the VA Pharmacy Benefits Management Service, NPOP has constructed user-friendly dashboards that allow users with no technical skills and who have the appropriate data access permissions to view various portions of the NPOP database. There are dashboards to display a list of NPOP samples by facility, find a patient by name or other identifying information, and display a list of patients who have received any antineoplastic drug, among other functions.
The NPOP database has been used to reannotate NGS results, such as when new drugs are approved. For example, when the first neurotrophic tropomyosin receptor kinase (NTRK) inhibitor was approved, NPOP rapidly identified all living patients with NTRK fusions and notified the health care providers of the availability a potential new treatment option for their patient. 7 NPOP is now building a method to allow NPOP dashboard users to similarly identify patients who have not received a corresponding drug for a user-selected LoE annotation. This will facilitate a systems approach to ensure that all patients with EGFR exon 19 deletions, for example, either have received an EGFR inhibitor or are reviewed to determine why they have not. Furthermore, the database will facilitate real-world data analysis in precision oncology. For example, prior to the recent FDA-approval of poly–(adenosine diphosphate–ribose) polymerase (PARP) inhibitors for prostate cancer, 50 veterans already had been treated with one of these agents. These data can help further inform which of the many variants of DNA damage repair genes benefit from PARP inhibitors.
Ensuring Access to Care for All Veterans
With any new medical technology comes an obligation to ensure appropriate equal access so as to not exacerbate health care disparities. Veterans enrolled in VA health care are much more likely to live in rural communities than does the US population as a whole, and there was concern that these veterans would not receive NGS testing at the same rate as urban veterans. NPOP therefore was intentional during implementation to ensure rural veterans were being offered testing. Indeed, there has been nearly equal utilization by rurality. No other disparities in NPOP utilization have been seen.
A majority of veterans who undergo testing in NPOP have at least 1 actionable gene variant reported.5 However, some of these are for lower LoE off-label use of FDA-approved medications, but many are for agents available only through clinical trials. Consideration of treatments available through a clinical trial is part of standard practice for patients with advanced malignancies. NPOP data have helped identify cohorts who are eligible for clinical trials on the basis of their tumor DNA sequencing results. The National Oncology Program Office has been working closely with the VA Office of Research and Development to expand access to cancer clinical trials in VA. Veterans also can be treated on trials outside VA as medically appropriate and with local VA approval.
Conclusions
The VA NPOP is one of the largest clinical DNA sequencing programs in the nation with integrated consultation services and health informatics resources to facilitate patient care, clinical trials, and health outcomes research. The clinical services of NPOP provide cuttingedge oncology services to veterans throughout VA without exacerbating disparities and will be a national resource for research.
Acknowledgments
NPOP was made possible and implemented through the efforts of a number of people in VHA, including the national and regional leaders who supported the program’s vision and implementation, especially Michael Mayo-Smith, David Shulkin, Jennifer S. Lee, and Laurence Meyer, the leaders and staff of the Massachusetts Veterans Epidemiology Research and Information Center who piloted regional NGS testing, and especially my current and former colleagues in the VA National Oncology Program Office, without whom NPOP would not be possible. The contributions of Neil L. Spector who served as inaugural Director of Precision Oncology and Jill E. Duffy in her role as Director of Oncology Operations are particularly noteworthy.
As the nation’s largest integrated health care system with about 50,000 new cancer diagnoses per year, providing care for over 400,000 veterans with cancer and a robust research portfolio, the US Department of Veterans Affairs (VA) is well positioned to be a leader in both clinical and research in oncology. The VA National Precision Oncology Program (NPOP), which provides tumor sequencing and consultative services, is a key component of VA oncology assets.
Case Presentation
As the mission of the VA is to “care for him who shall have borne the battle,” it is fitting to begin with the story of a US Army veteran in his 40s and the father of 2 young children who developed progressive shortness of breath, cough, and weight loss over a period of 8 months. He was diagnosed with metastatic lung adenocarcinoma in 2016, and standard testing of his tumor showed no alteration of the EGFR and ALK genes. He was treated with whole brain radiation and had begun treatment for carboplatin and pemetrexed chemotherapy with mixed tumor response.
Subsequently, his tumor was tested through NPOP, using a multigene next-generation sequencing (NGS) assay panel, which showed the presence of an abnormal fusion between the EML4 and ALK genes. The chemotherapy was discontinued and oral crizotinib precision therapy was started. The patient had an excellent response in all sites of disease (Figure 1). He was able to return to work and school.
In July 2017, his medication was switched to alectinib for asymptomatic progression in his brain, and there was further response. In September 2019, he was treated with precision intensity-modulated radiotherapy (IMRT), targeting a single brain metastasis as there were no other sites of cancer progression and no cancerrelated symptoms. He finished school and continues to work.
Precision Oncology
Oncology is a relatively young medical field. The early medical treatments for cancer were developed empirically against hematologic malignancies, particularly leukemias. Cytotoxic chemotherapeutic agents as a group have modest effects on most solid tumors, and even modern genomics has had limited ability to predict differential benefit in patients with advanced-stage carcinomas. As a result, the medications are used in a nonprecision manner in which all patients with the same cancer diagnosis and stage receive the same treatment. This is due in part to our limited understanding of both the pathophysiology of cancer and the mechanism of action of cytotoxic agents.
The paradigm of precision oncology, in contrast, utilizes unique, patient-specific molecular characteristics to guide prescribing of antineoplastic agents (Figure 2). These molecular characteristics are frequently tumoral but also may be nontumoral, such as germline genetic variants and even nonhuman, such as the gut microbiome as has been proposed as predictive of response to immune checkpoint inhibitors.1,2
One of the first examples of precision oncology was tumor testing for the estrogen receptor in breast cancer, which distinguishes breast tumors sensitive to hormonal treatments from those that are resistant.3 In 2004, somatically acquired mutation of the EGFR gene was found to be associated with response to EGFR tyrosine kinase inhibitors such as gefitinib and erlotinib, and subsequently it was shown that patients without these mutations derived no benefit from use of these drugs.4 Thus, the precision oncology paradigm is using a molecular diagnostic as part of the indication for an antineoplastic agent, resulting in improved therapeutic efficacy and often reduced toxicity.
By 2015, multiple examples of DNA-based gene alterations that predict drug response were known, including at least 5 in non-small cell lung cancer (NSCLC). The heterogeneity of molecular testing practice patterns and methods of testing in VA along with the increasing number and complexity of molecular tests facilitated launch of a regional precision oncology program based primarily in Veterans Integrated Service Network 1, which provided tumor DNA sequencing through 2 vendors. Advances in DNA sequencing technology, particularly NGS, permit sequencing of multiple genes in clinical tumor samples, using a panel applicable for multiple tumor types. As part of VA contributions to the 2016 White House Cancer Moonshot initiative, the regional program became NPOP with expanded geographic scope, the addition of clinical consultative services, and robust informatics that supports associated research and a learning health care system. NPOP is a component of the VA National Oncology Program Office under the Office of Specialty Care.
Testing
With the launch of NPOP in mid-2016, there was rapid expansion of the number of VA facilities participating, and the number of tumor samples being submitted increased substantially. 5 The expansion was facilitated by both central funding for the tumor DNA sequencing and by NPOP-provided training of pathology laboratory staff and oncologists. Today, NPOP is utilized by almost every oncology practice in VA.
NPOP’s initial focus was on lung cancer, specifically advanced-stage nonsquamous NSCLC, which not only is very common in VA, but also has one of the highest number of mutated genes that result in sensitivity to antineoplastic drugs. Recently, metastatic prostate cancer was added as a second focus tumor type. Dashboards are available on the NPOP website to assist care teams in identifying veterans at their facility with either lung or prostate cancer who may be appropriate for testing. Other solid tumors can be sent for testing through NPOP if patients have advanced stage cancer and are medically appropriate for antineoplastic therapy. To date, NPOP has sequenced > 13,000 samples.
Testing options have been added to NPOP in addition to tumor DNA sequencing. The first addition was the so-called liquid biopsy, more properly known as the cell-free DNA (cfDNA) test, a plasma-based high-sensitivity DNA sequencing assay. cfDNA is shed from dying cells and can be captured and sequenced from a plasma sample obtained by standard venipuncture, using a special-purpose sample collection tube. The test is appropriate for patients who do not have an appropriate archival tumor sample or those who cannot have a new biopsy of tumor tissue. Tumor tissue remains the preferred test sample due to a higher sensitivity than that of cfDNA and less susceptibility to false positives, so consideration of a tumor biopsy is appropriate prior to requesting a cfDNA assay. Therapy can greatly impact the sensitivity of cfDNA testing, so patients should be having disease progression at the time of obtaining a blood sample for cfDNA.
Finally, myeloid leukocytic cells accumulate genetic alterations during aging similar to those found in myelodysplasia and acute myeloid leukemia. These myeloid-associated mutations can be detected in both tumor and cfDNA samples and are known as clonal hyperplasia of indeterminate potential (CHIP). CHIP is much more common in the cfDNA. For lung cancer, CHIP-associated gene variants are readily distinguished from lung cancer-associated variants, but that distinction is much more difficult in many other tumor types.
In partnership with the current DNA sequencing contractor, NPOP provides access to a second gene panel for hematologic malignancies or sarcomas, though neither of these classes of malignancies currently have clear indications for routine NGS multigene panel testing. Given the low rate of finding a gene mutation that would change therapy that could not be found with smaller, less expensive gene panels, NPOP requires prior approval for the use of this panel.
Finally, since early 2019, programmed deathligand 1 (PD-L1) immunohistochemistry analysis is available through NPOP in association with NGS testing of the same sample for those solid tumors with US Food and Drug Administration (FDA)-approved indications that include a PD-L1 companion diagnostic. This service was added to facilitate concurrent testing of PD-L1 and DNA sequencing, which speeds availability of molecular data to the health care provider and veteran.
Determining Clinical Significance
The complexity of tumor NGS gene panel test results is far greater than frequently ordered laboratory or molecular testing due to the near infinite number of possible results and varying degrees of consensus of the significance of the results for therapeutic decision making. That complexity is reflected in the length of the test reports, which are often ≥ 20 pages. Starting from the gene variants identified by the DNA sequencing variant-caller bioinformatics pipeline, there is a 2-step process, referred to as annotation, to interpret the clinical significance that is repeated for each variant.
The first step is to assign a pathogenicity value, also known as oncogenicity, using a 5-point Likert scale from pathogenic to benign with variant of unknown significance (VUS) in the middle of the scale. Only variants that are pathogenic or likely pathogenic are considered further. A VUS is usually communicated to the health care provider but should generally not be acted on, while benign and likely benign variants may or may not be included in the report and should never be acted on. NPOP examined the concordance of pathogenicity calls among 3 annotation services: N-of-One/QCI Precision Insights (qiagen.com), IBM Watson for Genomics (WfG), and OncoKB (www.oncokb.org).6 There was moderate-to-poor concordance, indicating lack of consensus about whether a significant fraction of observed gene variants contributes to the patient’s cancer. This variability likely arises due to differences in algorithms and criteria used to assess pathogenicity.
The second step of annotation is assignment of the actionability of the variant, using a level of evidence (LoE) scale from 1 (on-label indication) to 4 (absence of clinical evidence; ie, only preclinical or theoretical evidence). Initially, NPOP used an adaptation of the LoE scales from WfG and OncoKB but now mostly uses the recently revised OncoKB LoE. Actionability also includes prediction of resistance to a treatment (LoE level R1 and R2). An example of a resistance gene variant is a KRAS mutation in colorectal cancer, which predicts lack of clinical benefit from anti- EGFR antibodies. It is important to note that a determination of actionability requires 3 inputs: gene, variant, and tumor type. A BRAF V600E mutation in melanoma has different medications with level 1 LoE than does the same mutation in colorectal cancer, for example.
Another complexity in annotation for actionability is tumor type ontogeny—the classification system used for cancer types. WfG uses a subset of the National Cancer Institute Thesaurus (ncithesaurus.nci.nih.gov), OncoKB uses the unique OncoTree (oncotree.mskcc.org), and Foundation Medicine (www.foundationmed icine.com), and N-of-One use propriety classification systems. The WfG and OncoKB tumor types have evolved over time, while it is unclear what changes have been made in the FMI and N-of-One tumor type classification systems. Thus, a gene variant observed in a single patient may be annotated differently by these services because of how the tumor type is mapped onto the services’ tumor type ontogeny. NPOP has been assigning WfG diagnoses since 2017, including historic assignment for prior samples back to the pilot project in 2015. In early 2019, NPOP began requiring test requesters to include International Classification of Diseases for Oncology, 3rd Edition (ICD-O-3) diagnoses (histology and site codes) with the sample. ICD-O-3 codes are used in all cancer registry data in North America, including the VA Cancer Registry System. The approximately 50,000 possible diagnoses allow fine precision in diagnoses, which is important for less common and rare cancer types; however, the large number of diagnoses adds complexity. NPOP has created a partial translation table for ICD-O-3 to WfG diagnosis that includes all diagnoses seen to date; this table facilitates continuing provision of WfG diagnosis without manual review as was previously required.
NPOP-Provided Genetic Services
Given these complexities in interpretation of NGS multigene panel results, NPOP provides several services to assist health care providers and other members of the care team. First, the NPOP Interfacility Consult (IFC) is a virtual “phone-a-friend” service that provides asynchronous patient-specific expert recommendations in precision oncology. By far the most requested service is assistance with interpretation of a patient’s DNA sequence results. Other requests include advice on whether to perform NGS testing and what molecular testing to perform. The IFC is integral to the VA Computerized Patient Record System electronic health record. Additional requests have been submitted and answered by e-mail.
The Molecular Oncology Tumor Board is a monthly case-based educational conference supported by the VA Employee Education Service, which provides continuing education credits for attendees. NPOP staff coordinate the conference, and a panel of specialists from around the country provide expert commentary.
In 2016, IBM gifted the services of WfG to VA. WfG’s main functionality is annotation of NGS results. About 5,000 samples were processed from 2017 to 2019; sample processing is expected to resume shortly. The availability of WfG annotations early in NPOP operation was very useful to the implementation of NPOP in general and the NPOP consultation services in particular, resulting in improved thoroughness of opinions provided by NPOP staff.
Informatics
Informatics is an essential component of NPOP that facilitates both clinical care and research (Figure 3). Results of NGS gene panels are returned to the facility that submitted the sample for testing as a PDF document. NPOP receives the same PDF report in real time but also structured data of the results including a variant callformat file and XML file. The secondary sequence data in binary alignment map or FASTQ format is received in batches. NPOP program staff extract data from these files and then load it into SQL tables in the VA Corporate Data Warehouse. In partnership with the VA Pharmacy Benefits Management Service, NPOP has constructed user-friendly dashboards that allow users with no technical skills and who have the appropriate data access permissions to view various portions of the NPOP database. There are dashboards to display a list of NPOP samples by facility, find a patient by name or other identifying information, and display a list of patients who have received any antineoplastic drug, among other functions.
The NPOP database has been used to reannotate NGS results, such as when new drugs are approved. For example, when the first neurotrophic tropomyosin receptor kinase (NTRK) inhibitor was approved, NPOP rapidly identified all living patients with NTRK fusions and notified the health care providers of the availability a potential new treatment option for their patient. 7 NPOP is now building a method to allow NPOP dashboard users to similarly identify patients who have not received a corresponding drug for a user-selected LoE annotation. This will facilitate a systems approach to ensure that all patients with EGFR exon 19 deletions, for example, either have received an EGFR inhibitor or are reviewed to determine why they have not. Furthermore, the database will facilitate real-world data analysis in precision oncology. For example, prior to the recent FDA-approval of poly–(adenosine diphosphate–ribose) polymerase (PARP) inhibitors for prostate cancer, 50 veterans already had been treated with one of these agents. These data can help further inform which of the many variants of DNA damage repair genes benefit from PARP inhibitors.
Ensuring Access to Care for All Veterans
With any new medical technology comes an obligation to ensure appropriate equal access so as to not exacerbate health care disparities. Veterans enrolled in VA health care are much more likely to live in rural communities than does the US population as a whole, and there was concern that these veterans would not receive NGS testing at the same rate as urban veterans. NPOP therefore was intentional during implementation to ensure rural veterans were being offered testing. Indeed, there has been nearly equal utilization by rurality. No other disparities in NPOP utilization have been seen.
A majority of veterans who undergo testing in NPOP have at least 1 actionable gene variant reported.5 However, some of these are for lower LoE off-label use of FDA-approved medications, but many are for agents available only through clinical trials. Consideration of treatments available through a clinical trial is part of standard practice for patients with advanced malignancies. NPOP data have helped identify cohorts who are eligible for clinical trials on the basis of their tumor DNA sequencing results. The National Oncology Program Office has been working closely with the VA Office of Research and Development to expand access to cancer clinical trials in VA. Veterans also can be treated on trials outside VA as medically appropriate and with local VA approval.
Conclusions
The VA NPOP is one of the largest clinical DNA sequencing programs in the nation with integrated consultation services and health informatics resources to facilitate patient care, clinical trials, and health outcomes research. The clinical services of NPOP provide cuttingedge oncology services to veterans throughout VA without exacerbating disparities and will be a national resource for research.
Acknowledgments
NPOP was made possible and implemented through the efforts of a number of people in VHA, including the national and regional leaders who supported the program’s vision and implementation, especially Michael Mayo-Smith, David Shulkin, Jennifer S. Lee, and Laurence Meyer, the leaders and staff of the Massachusetts Veterans Epidemiology Research and Information Center who piloted regional NGS testing, and especially my current and former colleagues in the VA National Oncology Program Office, without whom NPOP would not be possible. The contributions of Neil L. Spector who served as inaugural Director of Precision Oncology and Jill E. Duffy in her role as Director of Oncology Operations are particularly noteworthy.
1. Lima ZS, Ghadamzadeh M, Arashloo FT, Amjad G, Ebadi MR, Younesi L. Recent advances of therapeutic targets based on the molecular signature in breast cancer: genetic mutations and implications for current treatment paradigms. J Hematol Oncol. 2019;12(1):38. Published 2019 Apr 11. doi:10.1186/s13045-019-0725-6
2. Fessler J, Matson V, Gajewski TF. Exploring the emerging role of the microbiome in cancer immunotherapy. J Immunother Cancer. 2019;7(1):108. Published 2019 Apr 17. doi:10.1186/s40425-019-0574-4
3. Kiang DT, Kennedy BJ. Tamoxifen (antiestrogen) therapy in advanced breast cancer. Ann Intern Med. 1977;87(6):687- 690. doi:10.7326/0003-4819-87-6-687.
4. Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497-1500. doi:10.1126/science.1099314
5. Poonnen P, Duffy J, Hintze BJ, et al. Genomic analysis of metastatic solid tumors in veterans: findings from the VHA National Precision Oncology Program. J Clin Oncol. 2019;37(suppl 15):3074. doi:10.1200/JCO.2019.37.15_suppl.3074
6. Katsoulakis E, Duffy JE, Hintze B, Spector NL, Kelley MJ. Comparison of annotation services for nextgeneration sequencing in a large-scale precision oncology program. JCO Precis Oncol. 2020(4):212-221. doi:10.1200/PO.19.00118
7. Drilon A, Laetsch TW, Kummar S, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378(8):731-739. doi:10.1056/NEJMoa1714448
1. Lima ZS, Ghadamzadeh M, Arashloo FT, Amjad G, Ebadi MR, Younesi L. Recent advances of therapeutic targets based on the molecular signature in breast cancer: genetic mutations and implications for current treatment paradigms. J Hematol Oncol. 2019;12(1):38. Published 2019 Apr 11. doi:10.1186/s13045-019-0725-6
2. Fessler J, Matson V, Gajewski TF. Exploring the emerging role of the microbiome in cancer immunotherapy. J Immunother Cancer. 2019;7(1):108. Published 2019 Apr 17. doi:10.1186/s40425-019-0574-4
3. Kiang DT, Kennedy BJ. Tamoxifen (antiestrogen) therapy in advanced breast cancer. Ann Intern Med. 1977;87(6):687- 690. doi:10.7326/0003-4819-87-6-687.
4. Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497-1500. doi:10.1126/science.1099314
5. Poonnen P, Duffy J, Hintze BJ, et al. Genomic analysis of metastatic solid tumors in veterans: findings from the VHA National Precision Oncology Program. J Clin Oncol. 2019;37(suppl 15):3074. doi:10.1200/JCO.2019.37.15_suppl.3074
6. Katsoulakis E, Duffy JE, Hintze B, Spector NL, Kelley MJ. Comparison of annotation services for nextgeneration sequencing in a large-scale precision oncology program. JCO Precis Oncol. 2020(4):212-221. doi:10.1200/PO.19.00118
7. Drilon A, Laetsch TW, Kummar S, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378(8):731-739. doi:10.1056/NEJMoa1714448