User login
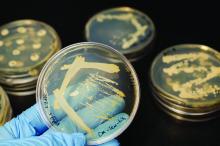
The first strain of Escherichia coli harboring the antibiotic-resistant genes mcr-1 and blaNDM-5 was isolated in the urine of a U.S. patient, according to a report in mBio.
A 76-year-old man was admitted to a tertiary-care hospital in New Jersey with a fever and flank pain in August 2014. The patient emigrated from India and resided in the United States for 1 year prior to this presentation. He had a history of prostate cancer treated with radiation therapy and subsequently developed recurrent urinary tract infections. He had also experienced bladder perforation requiring bilateral placement of nephrostomy tubes, which were clamped 5 days prior to presentation.
Using molecular analysis, the E. coli isolate from the study case (named MCR1_NJ) was shown to carry both mcr-1 and blaNDM-5 genes. In addition to mcr-1 and blaNDM-5, strain MCR1_NJ was found to harbor resistance genes for aminoglycosides, beta-lactams, chloramphenicol, fluoroquinolones, rifampin, sulfonamides, and tetracycline.
“This strain was isolated in August 2014, highlighting an earlier presence of mcr-1 within the region than previously known and raising the likelihood of ongoing undetected transmission,” wrote José R. Mediavilla, MBS, MPH of the New Jersey Medical School, Rutgers University, Newark, N.J., and his coauthors. “Active surveillance efforts involving all polymyxin- and carbapenem-resistant organisms are imperative in order to determine mcr-1 prevalence and prevent further dissemination.”
Find the full study in mBio (doi: 10.1128/mBio.01191-16).
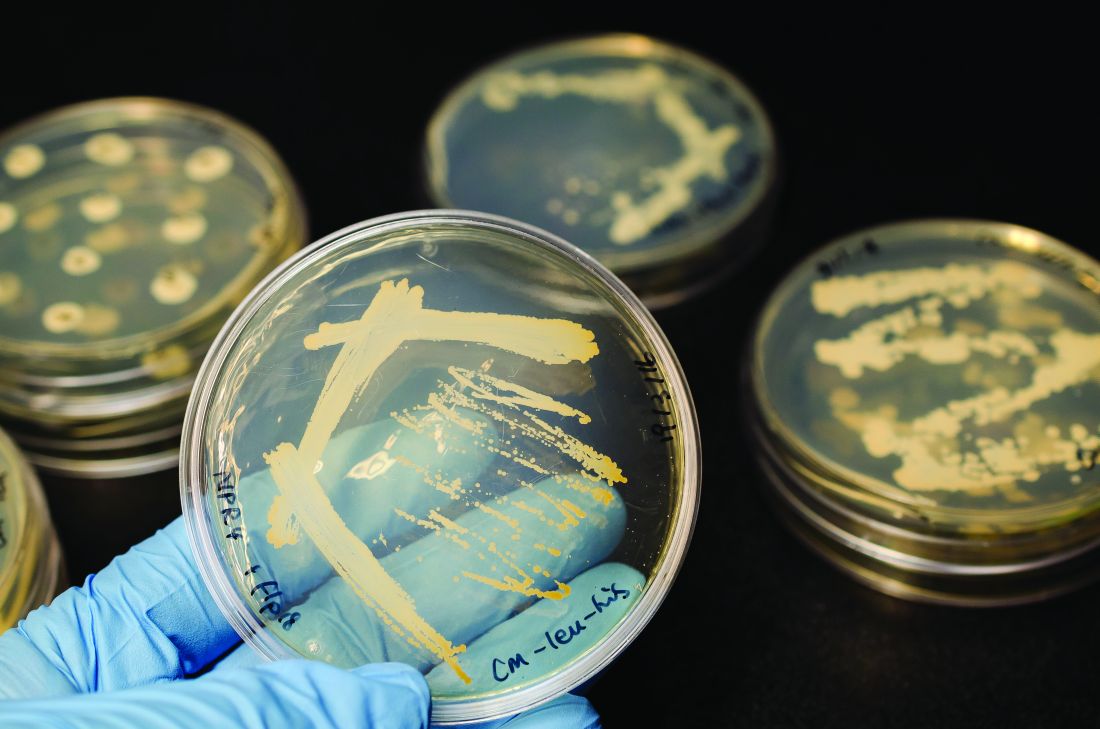
The first strain of Escherichia coli harboring the antibiotic-resistant genes mcr-1 and blaNDM-5 was isolated in the urine of a U.S. patient, according to a report in mBio.
A 76-year-old man was admitted to a tertiary-care hospital in New Jersey with a fever and flank pain in August 2014. The patient emigrated from India and resided in the United States for 1 year prior to this presentation. He had a history of prostate cancer treated with radiation therapy and subsequently developed recurrent urinary tract infections. He had also experienced bladder perforation requiring bilateral placement of nephrostomy tubes, which were clamped 5 days prior to presentation.
Using molecular analysis, the E. coli isolate from the study case (named MCR1_NJ) was shown to carry both mcr-1 and blaNDM-5 genes. In addition to mcr-1 and blaNDM-5, strain MCR1_NJ was found to harbor resistance genes for aminoglycosides, beta-lactams, chloramphenicol, fluoroquinolones, rifampin, sulfonamides, and tetracycline.
“This strain was isolated in August 2014, highlighting an earlier presence of mcr-1 within the region than previously known and raising the likelihood of ongoing undetected transmission,” wrote José R. Mediavilla, MBS, MPH of the New Jersey Medical School, Rutgers University, Newark, N.J., and his coauthors. “Active surveillance efforts involving all polymyxin- and carbapenem-resistant organisms are imperative in order to determine mcr-1 prevalence and prevent further dissemination.”
Find the full study in mBio (doi: 10.1128/mBio.01191-16).
The first strain of Escherichia coli harboring the antibiotic-resistant genes mcr-1 and blaNDM-5 was isolated in the urine of a U.S. patient, according to a report in mBio.
A 76-year-old man was admitted to a tertiary-care hospital in New Jersey with a fever and flank pain in August 2014. The patient emigrated from India and resided in the United States for 1 year prior to this presentation. He had a history of prostate cancer treated with radiation therapy and subsequently developed recurrent urinary tract infections. He had also experienced bladder perforation requiring bilateral placement of nephrostomy tubes, which were clamped 5 days prior to presentation.
Using molecular analysis, the E. coli isolate from the study case (named MCR1_NJ) was shown to carry both mcr-1 and blaNDM-5 genes. In addition to mcr-1 and blaNDM-5, strain MCR1_NJ was found to harbor resistance genes for aminoglycosides, beta-lactams, chloramphenicol, fluoroquinolones, rifampin, sulfonamides, and tetracycline.
“This strain was isolated in August 2014, highlighting an earlier presence of mcr-1 within the region than previously known and raising the likelihood of ongoing undetected transmission,” wrote José R. Mediavilla, MBS, MPH of the New Jersey Medical School, Rutgers University, Newark, N.J., and his coauthors. “Active surveillance efforts involving all polymyxin- and carbapenem-resistant organisms are imperative in order to determine mcr-1 prevalence and prevent further dissemination.”
Find the full study in mBio (doi: 10.1128/mBio.01191-16).