User login
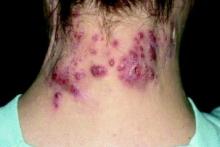
Hidradenitis suppurativa (HS) is a chronic inflammatory skin condition with lesions that include deep-seated nodules and abscesses, draining tracts, and fibrotic scars, and has been reported to be highly heritable.
A genomewide association study (GWAS) that involved a meta-analysis of data from three large biobanks (UK Biobank, FinnGen, and BioVU) identified one significant locus and two suggestive loci. The researchers were able to replicate the association with HS for two loci near the SOX9 and KLF5 genes in BioVU.
In addition, they also looked at genetic correlations between HS and autoimmune and inflammatory diseases, and results suggested a positive association with inflammatory bowel disease, psoriasis, and type 2 diabetes.
However, while plausible, the variant associations near candidate genes do not prove a causal effect of these variants or genes on disease risk, and further study is needed.
“It’s possible that these genes aren’t the ones affected by the variations we describe, but both are very strong candidates,” said study author Christopher J. Sayed, MD, associate professor of dermatology, University of North Carolina at Chapel Hill. “This improves our understanding of HS as a disease that is likely related to genetic changes that result in dysregulated hair follicle maintenance, inflammation, and wound healing.”
In turn, this will allow clinicians to educate patients about the underlying genetic influences on HS, he explained, as opposed to stigmatizing misconceptions focusing only on weight, smoking, and hygiene.
“Larger studies are underway and will be needed to find variants that may predict things like disease severity and response to treatment, but this is a big first step in the right direction,” Dr. Sayed added.
The study was published online in JAMA Dermatology.
Loci identified
GWASs have been limited in HS, and previous research has not identified significant risk loci. In the current study, Dr. Sayed and colleagues sought to identify underlying genes and genetic mechanisms that may underlie pathogenesis in HS.
They performed a GWAS in a cohort of 720 patients who were part of the Hidradenitis Suppurativa Program for Research and Care Excellence (HS ProCARE) at the UNC department of dermatology, and controls from the National Longitudinal Study of Adolescent to Adult Health (Add Health) study, a U.S.-based study following adolescents through adulthood.
The UK Biobank (UKB) is a prospective biobank with about 500,000 individuals aged 40-69 years, and FinnGen collects and analyzes genome and health data from Finnish biobank participants. To increase power to detect associations, a GWAS was performed using participants from the UKB (247 HS cases, 453,048 controls). The HS ProCARE GWAS results were meta-analyzed with UKB, along with data from FinnGen (673 HS cases, 297,544 controls). This three-way meta-analysis revealed one genomewide significant locus and two suggestive loci.
The authors found that the most strongly associated variant, rs10512572 located on chromosome 17, showed the strongest association in FinnGen; at the second locus, the most strongly associated variant was rs17090189 located on chromosome 13 and also showed the strongest association in FinnGen; and at the third locus, the most strongly associated variant, rs5792315, located on chromosome 11, showed the strongest association in HS ProCARE.
Next, they tested the 10 most strongly associated variants at the three loci in the BioVU biobank, which has 290 HS cases, including 189 individuals of European ancestry and 101 individuals of African ancestry, with 64,234 and 12,105 controls, respectively. The locus on chromosome 17 was replicated in BioVU in the same direction of effect, while one variant at the chromosome 13 locus showed nominal evidence of association in the same direction.
In a four-way meta-analysis of BioVU combined with the other three studies, the chromosome locus became more significant and the chromosome 13 locus exceeded the genomewide significance threshold. In contrast, the chromosome 11 locus was not replicated in BioVU (P = .27).
The authors pointed out that variants at these loci are located in keratinocyte regulatory elements near the genes SOX9 and KLF5, which play a role in skin and follicular inflammation, but have not previously been associated with HS pathogenesis.
Finally, they looked to see if there were shared genetic components between HS and autoimmune and inflammatory diseases. A nominally significant genetic correlation was observed between HS and inflammatory bowel disease (P = .04), psoriasis (P = .03), and type 2 diabetes (P = .04), although none reached significance.
Different manifestation with genetic variant
In a related study, researchers assessed the prevalence of the NCSTN:c.671_682del variant among individuals with HS in Malta, the island country in the Mediterranean, which has a high prevalence of HS and its associated risk factors, particularly obesity.
Led by Dillon Mintoff, MD, of the department of pathology at the University of Malta, Msida, they conducted a cross-sectional study of 113 adults with HS. In this group, 14 (12.39%) were found to be heterozygous for the NCSTN:c.671_682del variant. Individuals who had this variant were more likely to develop symptoms earlier and to manifest them in atypical skin sites, including the scalp, neck, torso, and antecubital fossae. Additionally, even though their symptoms weren’t more severe than those without the variant, patients with the variant were more likely to require treatment with biologic agents.
Studies move genetics in HS forward
Writing in an accompanying editorial, Atlas Khan, PhD, and Lynn Petukhova, PhD, from Columbia University, New York, noted that both of these HS genetic studies “set a solid foundation for future studies aimed at understanding the biological and clinical relevance of new HS genetic evidence.”
“Each study suggests a series of experiments that will allow us to gain new knowledge about HS,” they wrote, including coming closer to providing patients with a genetic diagnosis.
In addition, evidence from the GWAS paper suggests that with “larger HS GWASs we will be able to better prioritize drug-repurposing studies,” concluded the editorialists. “Both of these goals will help to improve health outcomes for patients with HS and their family members.”
Dr. Sayed reported grants and/or personal fees from AbbVie, Novartis, UCB, Incyte, InflaRx, Alumis, and ChemoCentryx outside the submitted work; and serving as a volunteer member of the board of the HS Foundation and member of the European HS Foundation. The study in Malta was funded by the Government of Malta’s Tertiary Education Scholarship Scheme and Institutional Funds from the University of Malta. Dr. Mintoff reported grants from the Government of Malta Ministry for Education, Sport, Youth, Research and Innovation during the conduct of the study. Dr. Khan reported receiving grants from the National Institute of Diabetes and Digestive Kidney Diseases. Dr. Petukhova reported receiving grants from the National Institute of Arthritis and Musculoskeletal and Skin Diseases, Columbia University’s Precision Medicine Initiative, Herbert Irving Comprehensive Cancer Center, and Data Science Institute.
Hidradenitis suppurativa (HS) is a chronic inflammatory skin condition with lesions that include deep-seated nodules and abscesses, draining tracts, and fibrotic scars, and has been reported to be highly heritable.
A genomewide association study (GWAS) that involved a meta-analysis of data from three large biobanks (UK Biobank, FinnGen, and BioVU) identified one significant locus and two suggestive loci. The researchers were able to replicate the association with HS for two loci near the SOX9 and KLF5 genes in BioVU.
In addition, they also looked at genetic correlations between HS and autoimmune and inflammatory diseases, and results suggested a positive association with inflammatory bowel disease, psoriasis, and type 2 diabetes.
However, while plausible, the variant associations near candidate genes do not prove a causal effect of these variants or genes on disease risk, and further study is needed.
“It’s possible that these genes aren’t the ones affected by the variations we describe, but both are very strong candidates,” said study author Christopher J. Sayed, MD, associate professor of dermatology, University of North Carolina at Chapel Hill. “This improves our understanding of HS as a disease that is likely related to genetic changes that result in dysregulated hair follicle maintenance, inflammation, and wound healing.”
In turn, this will allow clinicians to educate patients about the underlying genetic influences on HS, he explained, as opposed to stigmatizing misconceptions focusing only on weight, smoking, and hygiene.
“Larger studies are underway and will be needed to find variants that may predict things like disease severity and response to treatment, but this is a big first step in the right direction,” Dr. Sayed added.
The study was published online in JAMA Dermatology.
Loci identified
GWASs have been limited in HS, and previous research has not identified significant risk loci. In the current study, Dr. Sayed and colleagues sought to identify underlying genes and genetic mechanisms that may underlie pathogenesis in HS.
They performed a GWAS in a cohort of 720 patients who were part of the Hidradenitis Suppurativa Program for Research and Care Excellence (HS ProCARE) at the UNC department of dermatology, and controls from the National Longitudinal Study of Adolescent to Adult Health (Add Health) study, a U.S.-based study following adolescents through adulthood.
The UK Biobank (UKB) is a prospective biobank with about 500,000 individuals aged 40-69 years, and FinnGen collects and analyzes genome and health data from Finnish biobank participants. To increase power to detect associations, a GWAS was performed using participants from the UKB (247 HS cases, 453,048 controls). The HS ProCARE GWAS results were meta-analyzed with UKB, along with data from FinnGen (673 HS cases, 297,544 controls). This three-way meta-analysis revealed one genomewide significant locus and two suggestive loci.
The authors found that the most strongly associated variant, rs10512572 located on chromosome 17, showed the strongest association in FinnGen; at the second locus, the most strongly associated variant was rs17090189 located on chromosome 13 and also showed the strongest association in FinnGen; and at the third locus, the most strongly associated variant, rs5792315, located on chromosome 11, showed the strongest association in HS ProCARE.
Next, they tested the 10 most strongly associated variants at the three loci in the BioVU biobank, which has 290 HS cases, including 189 individuals of European ancestry and 101 individuals of African ancestry, with 64,234 and 12,105 controls, respectively. The locus on chromosome 17 was replicated in BioVU in the same direction of effect, while one variant at the chromosome 13 locus showed nominal evidence of association in the same direction.
In a four-way meta-analysis of BioVU combined with the other three studies, the chromosome locus became more significant and the chromosome 13 locus exceeded the genomewide significance threshold. In contrast, the chromosome 11 locus was not replicated in BioVU (P = .27).
The authors pointed out that variants at these loci are located in keratinocyte regulatory elements near the genes SOX9 and KLF5, which play a role in skin and follicular inflammation, but have not previously been associated with HS pathogenesis.
Finally, they looked to see if there were shared genetic components between HS and autoimmune and inflammatory diseases. A nominally significant genetic correlation was observed between HS and inflammatory bowel disease (P = .04), psoriasis (P = .03), and type 2 diabetes (P = .04), although none reached significance.
Different manifestation with genetic variant
In a related study, researchers assessed the prevalence of the NCSTN:c.671_682del variant among individuals with HS in Malta, the island country in the Mediterranean, which has a high prevalence of HS and its associated risk factors, particularly obesity.
Led by Dillon Mintoff, MD, of the department of pathology at the University of Malta, Msida, they conducted a cross-sectional study of 113 adults with HS. In this group, 14 (12.39%) were found to be heterozygous for the NCSTN:c.671_682del variant. Individuals who had this variant were more likely to develop symptoms earlier and to manifest them in atypical skin sites, including the scalp, neck, torso, and antecubital fossae. Additionally, even though their symptoms weren’t more severe than those without the variant, patients with the variant were more likely to require treatment with biologic agents.
Studies move genetics in HS forward
Writing in an accompanying editorial, Atlas Khan, PhD, and Lynn Petukhova, PhD, from Columbia University, New York, noted that both of these HS genetic studies “set a solid foundation for future studies aimed at understanding the biological and clinical relevance of new HS genetic evidence.”
“Each study suggests a series of experiments that will allow us to gain new knowledge about HS,” they wrote, including coming closer to providing patients with a genetic diagnosis.
In addition, evidence from the GWAS paper suggests that with “larger HS GWASs we will be able to better prioritize drug-repurposing studies,” concluded the editorialists. “Both of these goals will help to improve health outcomes for patients with HS and their family members.”
Dr. Sayed reported grants and/or personal fees from AbbVie, Novartis, UCB, Incyte, InflaRx, Alumis, and ChemoCentryx outside the submitted work; and serving as a volunteer member of the board of the HS Foundation and member of the European HS Foundation. The study in Malta was funded by the Government of Malta’s Tertiary Education Scholarship Scheme and Institutional Funds from the University of Malta. Dr. Mintoff reported grants from the Government of Malta Ministry for Education, Sport, Youth, Research and Innovation during the conduct of the study. Dr. Khan reported receiving grants from the National Institute of Diabetes and Digestive Kidney Diseases. Dr. Petukhova reported receiving grants from the National Institute of Arthritis and Musculoskeletal and Skin Diseases, Columbia University’s Precision Medicine Initiative, Herbert Irving Comprehensive Cancer Center, and Data Science Institute.
Hidradenitis suppurativa (HS) is a chronic inflammatory skin condition with lesions that include deep-seated nodules and abscesses, draining tracts, and fibrotic scars, and has been reported to be highly heritable.
A genomewide association study (GWAS) that involved a meta-analysis of data from three large biobanks (UK Biobank, FinnGen, and BioVU) identified one significant locus and two suggestive loci. The researchers were able to replicate the association with HS for two loci near the SOX9 and KLF5 genes in BioVU.
In addition, they also looked at genetic correlations between HS and autoimmune and inflammatory diseases, and results suggested a positive association with inflammatory bowel disease, psoriasis, and type 2 diabetes.
However, while plausible, the variant associations near candidate genes do not prove a causal effect of these variants or genes on disease risk, and further study is needed.
“It’s possible that these genes aren’t the ones affected by the variations we describe, but both are very strong candidates,” said study author Christopher J. Sayed, MD, associate professor of dermatology, University of North Carolina at Chapel Hill. “This improves our understanding of HS as a disease that is likely related to genetic changes that result in dysregulated hair follicle maintenance, inflammation, and wound healing.”
In turn, this will allow clinicians to educate patients about the underlying genetic influences on HS, he explained, as opposed to stigmatizing misconceptions focusing only on weight, smoking, and hygiene.
“Larger studies are underway and will be needed to find variants that may predict things like disease severity and response to treatment, but this is a big first step in the right direction,” Dr. Sayed added.
The study was published online in JAMA Dermatology.
Loci identified
GWASs have been limited in HS, and previous research has not identified significant risk loci. In the current study, Dr. Sayed and colleagues sought to identify underlying genes and genetic mechanisms that may underlie pathogenesis in HS.
They performed a GWAS in a cohort of 720 patients who were part of the Hidradenitis Suppurativa Program for Research and Care Excellence (HS ProCARE) at the UNC department of dermatology, and controls from the National Longitudinal Study of Adolescent to Adult Health (Add Health) study, a U.S.-based study following adolescents through adulthood.
The UK Biobank (UKB) is a prospective biobank with about 500,000 individuals aged 40-69 years, and FinnGen collects and analyzes genome and health data from Finnish biobank participants. To increase power to detect associations, a GWAS was performed using participants from the UKB (247 HS cases, 453,048 controls). The HS ProCARE GWAS results were meta-analyzed with UKB, along with data from FinnGen (673 HS cases, 297,544 controls). This three-way meta-analysis revealed one genomewide significant locus and two suggestive loci.
The authors found that the most strongly associated variant, rs10512572 located on chromosome 17, showed the strongest association in FinnGen; at the second locus, the most strongly associated variant was rs17090189 located on chromosome 13 and also showed the strongest association in FinnGen; and at the third locus, the most strongly associated variant, rs5792315, located on chromosome 11, showed the strongest association in HS ProCARE.
Next, they tested the 10 most strongly associated variants at the three loci in the BioVU biobank, which has 290 HS cases, including 189 individuals of European ancestry and 101 individuals of African ancestry, with 64,234 and 12,105 controls, respectively. The locus on chromosome 17 was replicated in BioVU in the same direction of effect, while one variant at the chromosome 13 locus showed nominal evidence of association in the same direction.
In a four-way meta-analysis of BioVU combined with the other three studies, the chromosome locus became more significant and the chromosome 13 locus exceeded the genomewide significance threshold. In contrast, the chromosome 11 locus was not replicated in BioVU (P = .27).
The authors pointed out that variants at these loci are located in keratinocyte regulatory elements near the genes SOX9 and KLF5, which play a role in skin and follicular inflammation, but have not previously been associated with HS pathogenesis.
Finally, they looked to see if there were shared genetic components between HS and autoimmune and inflammatory diseases. A nominally significant genetic correlation was observed between HS and inflammatory bowel disease (P = .04), psoriasis (P = .03), and type 2 diabetes (P = .04), although none reached significance.
Different manifestation with genetic variant
In a related study, researchers assessed the prevalence of the NCSTN:c.671_682del variant among individuals with HS in Malta, the island country in the Mediterranean, which has a high prevalence of HS and its associated risk factors, particularly obesity.
Led by Dillon Mintoff, MD, of the department of pathology at the University of Malta, Msida, they conducted a cross-sectional study of 113 adults with HS. In this group, 14 (12.39%) were found to be heterozygous for the NCSTN:c.671_682del variant. Individuals who had this variant were more likely to develop symptoms earlier and to manifest them in atypical skin sites, including the scalp, neck, torso, and antecubital fossae. Additionally, even though their symptoms weren’t more severe than those without the variant, patients with the variant were more likely to require treatment with biologic agents.
Studies move genetics in HS forward
Writing in an accompanying editorial, Atlas Khan, PhD, and Lynn Petukhova, PhD, from Columbia University, New York, noted that both of these HS genetic studies “set a solid foundation for future studies aimed at understanding the biological and clinical relevance of new HS genetic evidence.”
“Each study suggests a series of experiments that will allow us to gain new knowledge about HS,” they wrote, including coming closer to providing patients with a genetic diagnosis.
In addition, evidence from the GWAS paper suggests that with “larger HS GWASs we will be able to better prioritize drug-repurposing studies,” concluded the editorialists. “Both of these goals will help to improve health outcomes for patients with HS and their family members.”
Dr. Sayed reported grants and/or personal fees from AbbVie, Novartis, UCB, Incyte, InflaRx, Alumis, and ChemoCentryx outside the submitted work; and serving as a volunteer member of the board of the HS Foundation and member of the European HS Foundation. The study in Malta was funded by the Government of Malta’s Tertiary Education Scholarship Scheme and Institutional Funds from the University of Malta. Dr. Mintoff reported grants from the Government of Malta Ministry for Education, Sport, Youth, Research and Innovation during the conduct of the study. Dr. Khan reported receiving grants from the National Institute of Diabetes and Digestive Kidney Diseases. Dr. Petukhova reported receiving grants from the National Institute of Arthritis and Musculoskeletal and Skin Diseases, Columbia University’s Precision Medicine Initiative, Herbert Irving Comprehensive Cancer Center, and Data Science Institute.
FROM JAMA DERMATOLOGY