User login
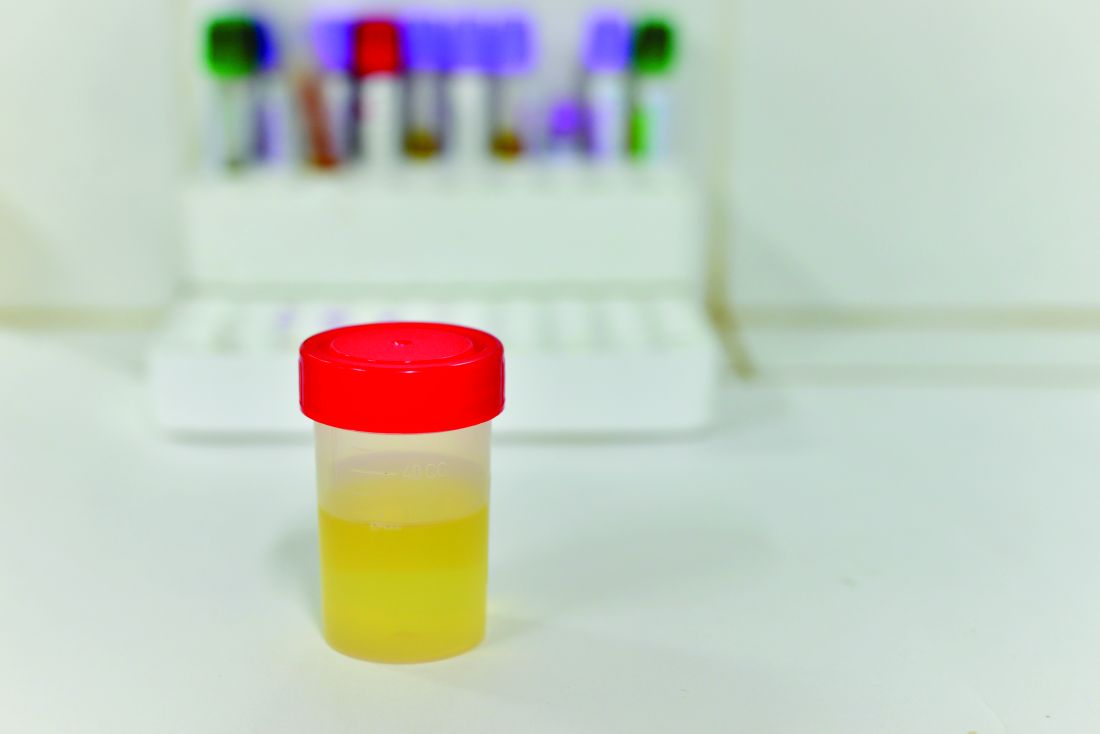
Researchers in Sweden have developed a 30-minute test capable of determining whether a bacterial urinary tract infection is susceptible or resistant to nine antibiotics. Their findings suggest that it is possible to develop a point-of-care test for patients with UTI.
Most phenotypic and genotypic antibiotic susceptibility tests are too slow to guide treatment, ranging from 2 days to 1 hour. The researchers at Uppsala (Sweden) University cut the testing time down to less than 30 minutes by using a microfluidic chip and direct single-cell imaging.
The chip traps the bacterial cells and allows growth media with different antibiotics (or none) to flow around them. “With this setup, we could detect the differential growth rate between treatment and reference populations in 3 min for ciprofloxacin, levofloxacin, mecillinam, nitrofurantoin, and trimethoprim-sulfamethoxazole; 7 min for amoxicillin-clavulanate and doripenem; 9 min for fosfomycin; and 11 min for ampicillin based on 99.9% confidence intervals,” wrote Özden Baltekin and his coauthors.
That test specifically used Escherichia coli cells; comparable speed and accuracy was replicated using Klebsiella pneumoniae and Staphylococcus saprophyticus. For the development of a point-of-care test for patients, the researchers said all that would be needed are about 100 bacteria cells.
“We have here focused on bacterial species and antibiotics related to UTIs, but it is likely that the same principles would work for sepsis, mastitis, or meningitis,” they suggested (Proc Natl Acad Sci. 2017 Aug 8. doi: 10.1073/pnas.1708558114).
Researchers in Sweden have developed a 30-minute test capable of determining whether a bacterial urinary tract infection is susceptible or resistant to nine antibiotics. Their findings suggest that it is possible to develop a point-of-care test for patients with UTI.
Most phenotypic and genotypic antibiotic susceptibility tests are too slow to guide treatment, ranging from 2 days to 1 hour. The researchers at Uppsala (Sweden) University cut the testing time down to less than 30 minutes by using a microfluidic chip and direct single-cell imaging.
The chip traps the bacterial cells and allows growth media with different antibiotics (or none) to flow around them. “With this setup, we could detect the differential growth rate between treatment and reference populations in 3 min for ciprofloxacin, levofloxacin, mecillinam, nitrofurantoin, and trimethoprim-sulfamethoxazole; 7 min for amoxicillin-clavulanate and doripenem; 9 min for fosfomycin; and 11 min for ampicillin based on 99.9% confidence intervals,” wrote Özden Baltekin and his coauthors.
That test specifically used Escherichia coli cells; comparable speed and accuracy was replicated using Klebsiella pneumoniae and Staphylococcus saprophyticus. For the development of a point-of-care test for patients, the researchers said all that would be needed are about 100 bacteria cells.
“We have here focused on bacterial species and antibiotics related to UTIs, but it is likely that the same principles would work for sepsis, mastitis, or meningitis,” they suggested (Proc Natl Acad Sci. 2017 Aug 8. doi: 10.1073/pnas.1708558114).
Researchers in Sweden have developed a 30-minute test capable of determining whether a bacterial urinary tract infection is susceptible or resistant to nine antibiotics. Their findings suggest that it is possible to develop a point-of-care test for patients with UTI.
Most phenotypic and genotypic antibiotic susceptibility tests are too slow to guide treatment, ranging from 2 days to 1 hour. The researchers at Uppsala (Sweden) University cut the testing time down to less than 30 minutes by using a microfluidic chip and direct single-cell imaging.
The chip traps the bacterial cells and allows growth media with different antibiotics (or none) to flow around them. “With this setup, we could detect the differential growth rate between treatment and reference populations in 3 min for ciprofloxacin, levofloxacin, mecillinam, nitrofurantoin, and trimethoprim-sulfamethoxazole; 7 min for amoxicillin-clavulanate and doripenem; 9 min for fosfomycin; and 11 min for ampicillin based on 99.9% confidence intervals,” wrote Özden Baltekin and his coauthors.
That test specifically used Escherichia coli cells; comparable speed and accuracy was replicated using Klebsiella pneumoniae and Staphylococcus saprophyticus. For the development of a point-of-care test for patients, the researchers said all that would be needed are about 100 bacteria cells.
“We have here focused on bacterial species and antibiotics related to UTIs, but it is likely that the same principles would work for sepsis, mastitis, or meningitis,” they suggested (Proc Natl Acad Sci. 2017 Aug 8. doi: 10.1073/pnas.1708558114).
FROM PNAS