User login
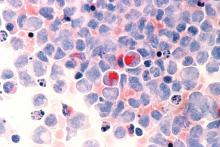
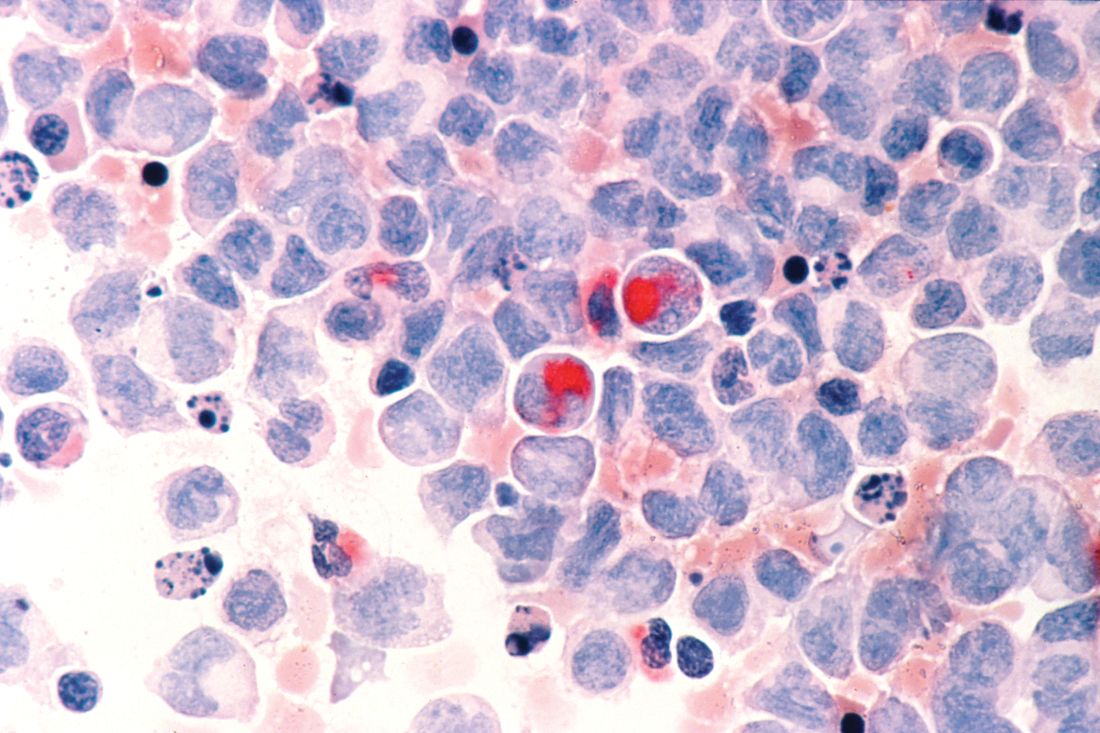
Researchers in the United States and the United Kingdom believe they have found a new gene target that could aid in the development of more effective treatments for acute myeloid leukemia (AML).
Inhibition of the METTL3 gene allowed for the destruction of AML in human and mouse cells without damaging healthy blood cells, the researchers reported in a research letter published Nov. 27 in the journal Nature.
Using mouse cells, the researchers used CRISPR-Cas9 gene editing technology to identify RNA-modifying enzymes that are needed for the survival and proliferation of AML cells. They identified 46 potential candidate genes and further narrowed that to the METTL gene families. They next targeted METTL1, METTL3, METTL14, and METTL16 in 10 human AML cell lines and 10 cell lines from heterogeneous cancer types. METTL3 was shown to have the strongest effect.
Read the full research letter in Nature (doi: 10.1038/nature24678).
mschneider@frontlinemedcom.com
On Twitter @maryellenny
Researchers in the United States and the United Kingdom believe they have found a new gene target that could aid in the development of more effective treatments for acute myeloid leukemia (AML).
Inhibition of the METTL3 gene allowed for the destruction of AML in human and mouse cells without damaging healthy blood cells, the researchers reported in a research letter published Nov. 27 in the journal Nature.
Using mouse cells, the researchers used CRISPR-Cas9 gene editing technology to identify RNA-modifying enzymes that are needed for the survival and proliferation of AML cells. They identified 46 potential candidate genes and further narrowed that to the METTL gene families. They next targeted METTL1, METTL3, METTL14, and METTL16 in 10 human AML cell lines and 10 cell lines from heterogeneous cancer types. METTL3 was shown to have the strongest effect.
Read the full research letter in Nature (doi: 10.1038/nature24678).
mschneider@frontlinemedcom.com
On Twitter @maryellenny
Researchers in the United States and the United Kingdom believe they have found a new gene target that could aid in the development of more effective treatments for acute myeloid leukemia (AML).
Inhibition of the METTL3 gene allowed for the destruction of AML in human and mouse cells without damaging healthy blood cells, the researchers reported in a research letter published Nov. 27 in the journal Nature.
Using mouse cells, the researchers used CRISPR-Cas9 gene editing technology to identify RNA-modifying enzymes that are needed for the survival and proliferation of AML cells. They identified 46 potential candidate genes and further narrowed that to the METTL gene families. They next targeted METTL1, METTL3, METTL14, and METTL16 in 10 human AML cell lines and 10 cell lines from heterogeneous cancer types. METTL3 was shown to have the strongest effect.
Read the full research letter in Nature (doi: 10.1038/nature24678).
mschneider@frontlinemedcom.com
On Twitter @maryellenny
FROM NATURE