User login
COVID-19 Cycle Threshold/Cycle Number Testing at a Community Living Center
COVID-19, caused by SARS-CoV-2, is more severe in individuals with underlying illnesses. Because complete social distancing might be more difficult in nursing homes and community living centers (CLCs), public health leaders and clinicians have been concerned about the epidemiology and disease course in nursing homes even before the COVID-19 pandemic.1-7 A report of a COVID-19 outbreak in a nursing home facility in King County, Washington, documented a 33.7% overall fatality rate for residents and 52.4% among the most critically ill.4,5 The experience at King County, Washington, shows that proactive steps to identify, monitor, and apply preventive control measures is important for future outbreaks.5
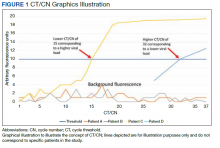
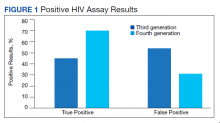
Reverse transcriptase polymerase chain reaction (RT-PCR) testing produces a cycle threshold (CT) or cycle number (CN) that correlates with viral load and infectiousness. 8-14 CT/CN represents the number of RT-PCR cycles required for the fluorescent signal to cross the detection threshold (exceed background level) and is inversely proportional to the viral load. Effectively, the higher the viral load, the lower the CT/ CN value (Figure 1). Tracking CT/CN values was not documented in the Washington nursing home outbreak. Reports of COVID- 19 testing in CLCs during outbreaks are sparse, and CT/CN values and demographic distribution of these veterans has not been reported.15 The CLC veteran population, with known higher vulnerability to infection and chronic diseases, is epidemiologically different from the general nursing home population.15-18 To address these literature gaps, we present the first report of COVID- 19 testing with CT/CN value correlations in the high-risk veteran CLC population.
Methods
A retrospective review of all COVID-19 CT/CN testing at the Corporal Michael J. Crescenz Veterans Affairs Medical Center (VAMC) CLC in Philadelphia, Pennsylvania, from March 28, 2020, to April 24, 2020, was performed with a US Department of Veterans Affairs (VA) Veterans Health Information System Architecture VistA/FileMan search. Only veteran residents were included in this review. Data collected included initial and serial test results, CT/CN on positive test results, test dates, testing platform used, demographic information (age, self-reported ethnicity, and sex), and clinical follow-up information. Health records were reviewed retrospectively to identify death, the first day after diagnosis with no documented symptoms, or hospitalization status.
RT-PCR testing was performed with the Abbott RealTime SARS-CoV-2 assay on the Abbott m2000 platform and the Xpert Xpress SARS-CoV-2 assay on the Cepheid Infinity platform. The Xpert Xpress assay gave 2 CT values for the E and N2 targets on positive samples.19 For this assay to indicate a positive specimen, amplification by RT-PCR of the N2 target or both the N2 and E target is required. The Xpert Xpress assay results as presumptive positive if only the E target amplified. This assay counts a maximum of 45 cycles. The Abbott RealTime SARS-CoV-2 assay gave 1 CN derived from the RNA-dependent RNA polymerase and N targets on positive samples.20 The Abbott assay on the m2000 counts a maximum of 37 cycles. The CT/CN value is the number of cycles required by RT-PCR for the fluorescence signal to cross a threshold value exceeding background level.19,20
Samples that are negative for COVID-19 by RT-PCR do not produce a CT/CN value. Although both instruments were used for RT-PCR, the precise CT/CN values are not interchangeable and CT/CN observations over time between the 2 instruments during the disease course would be based on CT/CN value movement (general upward or downward trend) rather than absolute CT/CN differences. Both assays have been approved by emergency use authorization as qualitative tests for the presence/absence of COVID-19. Although the CT/CN value is available to laboratory staff after test completion, the CT/CN value is not reported routinely in the patient health record. All veteran patients identified on the initial review from March 28, 2020, to April 24, 2020, had all serial COVID-19 testing recorded until November 10, 2020. The CN values at the limit of detection (LOD) for the Abbott m2000 platform from the initial validation study were reviewed for reference.21
Results
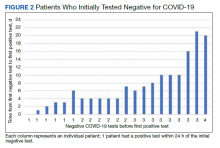
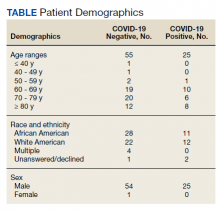
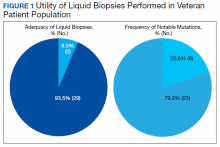
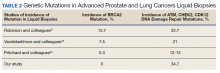
Of 80 patients, 25 (31%) were COVID-19 positive over the course of testing. The study population had a mean age of 73.5 years; 92% were aged > 60 years. The group was predominantly male (79 male vs 1 female). Among the 77 patients with a stated ethnicity, 39 (51%) were African American. In comparison, 43% of residents in Philadelphia County are African American (Table).22,23 Additionally, a previously published total COVID-19 tested population by ethnicity at the same regional VAMC revealed 46.8% of tested veteran patients were African American. 24 Three patients had no stated ethnicity. Among those who tested positive, 11 were African American patients, 12 were White patients, and 2 had no stated ethnicity. Four patients tested positive on their first test. The other 21 patients were positive on repeat testing. Interestingly, 6 patients had 1 initial negative test before a positive test, 6 patients had 2, 8 patients had 3, and 1 patient had 4 initial negative tests before a positive test result. Among the 25 positive patients, 22 were either positive within 10 days of the initial negative test result or initially positive (Figure 2). Three patients who tested positive after 10 days did so at 16, 20, and 21 days after the initial negative test result. Among the 25 positive patients, 23 had initial and serial testing from both the Abbott and Xpert Xpress assays. The remaining 2 positive patients had initial and serial testing from the Abbott assay exclusively.
Only positive COVID-19 results by RTPCR produced a CT/CN value. After disease resolution with a negative test, no CT/CN value was produced with the negative test result on either testing platform. Because repeat testing after the initial positive result took place no sooner than 10 days, we observed that the CT/CN value increased after the initial positive result until the disease resolved, and a negative result was obtained (eAppendix 1, available online at doi:10.12788/fp.0276). A t test comparing the initial CT/CN value to the value more than 10 days after the initial positive showed the CT/CN was statistically significantly higher (P < .05).
Prompt repeat testing after the initial test can show a decrease in the CT/CN value because of increasing viral load before the expected increase until disease resolution if the initial test caught the infection early. Twelve patients had a negative test result between 2 serial positive results. These negative test results occurred later, near the end of the disease course. Among the 12 patients with this positive-negativepositive CT/CN pattern, 7 were symptomatic and no longer had documented symptoms or hospitalization around the time of this positive-negative-positive pattern. Four of these individuals were asymptomatic during the entire infection course. One of the 12 patients with this pattern expired with the negative result occurring on day 27 of the disease in the context of rising CT/CN. One of these 12 patients only had a presumptive positive test result on the Cepheid because it detected only the E target with a CT value of 38.7. In 1 of the 12 patients, the negative test result occurred between 2 positive test results with CT/CN values < 20 (12.05 and 19.05 for the positive tests before and after the negative result, respectively). When the initial CT/CN values was separated based on ethnicity, the average CT/CN value for African Americans (23.3) was higher than for other ethnicities (19.9), although it did not reach statistical significance (P = .35).
Ten of the 25 patients testing positive were admitted to the hospital, including 1 admitted 15 days before diagnosis (patient 20) and 1 admitted 80 days after diagnosis (patient 7). Among these 10 patients, 6 were admitted to the intensive care unit, including patient 7. None of the patients were intubated. Three of the 10 admitted patients died (patients 7, 20, and 24). Patient 7 was a 79-year-old male with a history of dementia, cerebrovascular accident, hypertension, hyperlipidemia, and chronic kidney disease with symptoms of lethargy and refusal of oral intake when he was diagnosed with COVID-19. He was admitted 80 days after diagnosis for hyponatremia and acute renal failure, with death on day 87 recorded as complications from the earlier COVID-19 infection. Patient 20, an 89-year-old male with a history of dementia, chronic kidney disease, and hyperlipidemia, had been admitted with fever, cough, and leukocytosis 17 days before COVID-19 diagnosis. He continued to be symptomatic after diagnosis with development of hypotension, dehydration, and refusal of oral intake while on comfort measures/endof- life care and died 15 days after COVID- 19 infection diagnosis. Patient 24 was a 96-year-old male with history of heart failure, hypertension, coronary artery disease, prostate carcinoma, and dementia who developed a cough at the time of diagnosis; because of his underlying condition, he remained in the CLC on comfort care. His symptoms, including hypoxia, worsened until he died 7 days after diagnosis.
Among the 25 patients, 17 were symptomatic at the time of diagnosis; the 14 initially symptomatic patients who survived improved clinically and returned to baseline. Eight of the 25 patients were asymptomatic initially and 3 developed symptoms 2 to 5 days after diagnosis. Only 1 patient who remained asymptomatic was admitted for inability to adhere to quarantine at the CLC. Review of the health records of all surviving symptomatic patients showed symptom resolution with return to baseline that corresponds to an increasing CT/CN value. A 1-tailed t test comparing the initial CT/ CN at the time of diagnosis to the last CT/CN value for symptomatic patients who recovered revealed a statistically significant increase (P < .05). For the symptomatic, symptom resolution and hospital discharge took (if required) a mean 20 days (range, 7-46). Among those who were not hospitalized, symptoms resolved in 7 to 36 days (18 days). Among those requiring hospitalization at any time (excluding patients who died or were asymptomatic), symptom and hospitalization resolution took a mean 22 days (range, 10-46). Asymptomatic patients (patients 8, 10, 15, 16, and 25) also showed increasing CT/CN value during the infection course, although there was no correlation with the continued lack of symptoms.
During the initial validation of the Abbott m2000 instrument, an LOD study included concentrations of 1000, 500, 250, 100, 70, 60, and 50 virus copies/mL (eAppendix 2, available online at doi:10.12788/fp.0276).21 The average CN at 100 virus copies/mL—the manufacturer provided LOD in the instructions for use—was 25.74.20 At a concentration of one-half that (50 virus copies/mL), the average CN was 28.39.
Discussion
This is the first study in the English literature to track CT/CN values as part of serial testing of a veteran CLC. Widescale testing and repeat screening in the absence of symptoms of nursing home residents would identify those who are infected and allow providers to track viral load clearance.9-14 CT/CN values, when serially tracked during the infection course, appear to increase with illness resolution, consistent with earlier reports that CT/CN correlates with viral load.8-14 Serial CT/CN values that are high (> 25) and continue to increase with each test suggest progression toward disease resolution or viral RNA clearance.8-14 After symptom resolution, patients can have a persistent low level of viral shedding (corresponding to a high CT/CN value).10-14,25 Near the end of disease resolution, a negative serial RT-PCR sample test before a subsequent positive might be a promising clinical sign of near disease recovery. Once the viral load is low with a CT/CN significantly higher than 25, some specimens might result as negative but turn up positive on subsequent sampling with a high CT/CN value. This pattern, with attendant high CT/CN values for the positive results, are consistent with the known effect of viral load (ie, a low viral load correlates to a high CT/CN) and adequacy of specimen collection on CT/CN values.25 If the patient’s viral load is low, the sample collected might have a viral load at or near the testing platform’s LOD.
For Abbott m2000, the manufacturer provided LOD is 100 virus copies/mL, although the instrument was able to detect virus concentrations below that level during the initial validation.20 The actual LOD of the instrument at our institution is < 100 virus copies/mL. For the Cepheid Xpert Xpress SARS-CoV-2 assay, the manufacturer-provided LOD is 250 virus copies/mL.19 An LOD study including samples below the manufacturer-provided LOD was not part of the initial validation study for the Xpert Xpress assay. Nonetheless, the virus concentration of samples with very high CT values at or near the maximum CT value of 45 is expected to be at or near the platform’s actual LOD.
If the samples collected near the end of the patient’s disease course have viral loads near these low concentrations, the encouraging positive-negative-positive pattern with high CT/CN values might be a promising sign for viral clearance. On the other hand, a positive-negative-positive pattern in the setting of low CT/CN values before and after the negative test might indicate poor sampling for the negative specimen. The back-and-forth or positive-negative-positive pattern generally appears to indicate near resolution of the infection course, although clinical correlation is necessary to rule out inadequate sampling earlier in the disease course or prolonged viral RNA shedding.9-14 In all of the surviving symptomatic patients who showed the positive-negative-positive pattern, this sign occurred around or after symptom resolution. It also is important to consider that in some patients, SARS-CoV-2 RNA might remain detectable with increasing CT/CN after symptom resolution, and samples from these patients might not result positive. Therefore, CT/CN values cannot be interpreted without considering the clinical picture.25
Studies on infectiousness and virus culture from COVID-19 samples with CT/ CN correlation have shown that patients with high CT/CN at the end of their disease course might not be as infectious.9-14,25 Because 1 patient had a presumptive positive result after the negative result, this study shows that this positive-negative-positive pattern could include presumptive positive results. Also, in the setting of a recent positive result on the same testing platform, a patient with this pattern is presumed to be positive for COVID-19 RNA because of scant viral material.
Taiwan’s public health response to the outbreak illustrates the ability to mitigate an outbreak throughout a society.26 These actions could help blunt an outbreak within a civilian nursing home population.5 Mitigation within a veteran CLC population has been documented, but the study, which focused on mitigation, did not consider CT/CN values, demographic distribution, testing access of the studied population, or laboratory findings related to disease pathophysiology.15 A key ingredient in widescale, serial testing is the availability of a rapid turnaround from testing in-house that allowed identification within 24 hours instead of several days at a reference laboratory. 15 Rapid widescale testing would allow clinical teams to optimize the Triangle of Benefit of Widescale Timely Tests for CLC (Figure 3).15 Timely laboratory testing remains pivotal for CLC veteran residents to aid successful clinical triage and management. Reporting serial CT/CN values can provide additional information to clinicians about the disease course because CT/ CN correlates with viral load, which varies based on where the patient is in the disease course.9-14 CT/CN values carry significant prognostic value, particularly with respect to intubation and mortality.8
Limitations
Important limitations to our study include the use of 2 separate RT-PCR platforms. Using different RT-PCR platforms is common in clinical laboratories trying to take advantage of the unique characteristics of different platforms—for example, turnaround time vs high throughput— to manage COVID-19 testing workflow.25 However, the exact CT/CN values obtained from each platform might not translate to the other, and the general trend (CT/CN values are rising or falling across serial tests) rather than a single value could be useful for clinical correlation. Even when the same platform is used for the serial testing, CT/CN values can be affected by adequacy of specimen collection; therefore, clinical correlation and considering the trend in CT/CN values is necessary for interpretation.10-14,25 Because of the known trend in viral dynamics, a positive specimen collected with a high CT/CN followed by a subsequent (within 2 days) positive specimen collected with a low CT/CN might be compatible with early detection of COVID- 19 infection in the appropriate clinical context. 10-14 However, detection late in the infection course or even after the symptomatic disease resolved with prolonged viral shedding might show serial positive samples with increasing CT/CN values.10-14
Patients with prolonged viral shedding might not be infectious.27 Because of the clinical correlation required for interpretation and the other factors that might affect CT/CN values, recommendations advise against using CT/CN values in clinical practice at this time, although these recommendations could change with future research.25 Serial CT/CN values have the potential, if appropriately correlated with the clinical picture, to provide useful information, such as whether the viral load of the sample is relatively high or low and increasing or decreasing.
Veterans, as a population, are more susceptible to poor health outcomes and morbidity compared with similar civilian counterparts.2,14-16 Veteran CLC patients likely would experience worse outcomes with COVID-19, including more infections, expiration, and morbidity compared with similar general population nursing homes. Similar to what had been reported for the civilian population, a trend (high CT/CN values early in the disease course with repeat testing needed to detect all positives followed by lower CT/CN value to correlate with increased viral load and then increased CT/CN value as the infection resolved) also was observed in this veteran population.
It has been extensively documented that minority groups experience decreased health care access and worse health outcomes. 28-30 Considering the critical medical supply shortages, including personal protective equipment, ventilators, and even testing supplies, there is the potential for a resource access disparity by ethnicity.28-31 Because the VA does not depend on measures of wealth and privilege such as health insurance, there was no disparity noted in access to testing by race or ethnicity at the VAMC CLC. When considering the health outcome of viral load from the measured CT/CN value, the viral loads of African American patients and those of other ethnicities was not significantly different in this study.
Conclusions
This is the first study to bring up critical points including serial CT/CN value correlation in RT-PCR tests, demographic distributions demonstrating easy and equal access in a veteran nursing home to COVID-19 testing, and clinical laboratory signs related to disease pathophysiology. Unlike other populations who have undergone serial CT/CN monitoring, nursing homes represent a particularly vulnerable population who require measures to prevent the spread and mitigate outbreaks of COVID-19.2,4,5 Test measurements obtained such as the CT/CN value during routine clinical care can provide useful information for public health, epidemiologic, or clinical purposes with appropriate correlation to clinical and other laboratory parameters. This study demonstrates early intervention of serial testing of an outbreak in a veterans nursing home with CT/CN value correlation.
1. Chen T, Wu D, Chen H, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. doi:10.1136/bmj.m1091
2. Tsan L, Davis C, Langberg R, et al. Prevalence of nursing home-associated infections in the Department of Veterans Affairs nursing home care units. Am J Infect Control. 2008;36(3):173-179. doi:10.1016/j.ajic.2007.06.008
3. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054-1062. doi:10.1016/S0140-6736(20)30566-3
4. Arentz M, Yim E, Klaff L, et al. Characteristics and outcomes of 21 critically ill patients with COVID-19 in Washington State. JAMA. 2020;323(16):1612-1614. doi:10.1001/jama.2020.4326
5. McMichael TM, Currie DW, Clark S, et al. Public Health–Seattle and King County, EvergreenHealth, and CDC COVID-19 Investigation Team. Epidemiology of Covid-19 in a long-term care facility in King County, Washington. N Engl J Med. 2020;382(21):2005-2011. doi:10.1056/NEJMoa2005412
6. Childs A, Zullo AR, Joyce NR, et al. The burden of respiratory infections among older adults in long-term care: a systematic review. BMC Geriatr. 2019;19(1):210. doi:10.1186/s12877-019-1236-6
7. Eriksen HM, Iversen BG, Aavitsland PJ. Prevalence of nosocomial infections and use of antibiotics in long-term care facilities in Norway, 2002 and 2003. Hosp Infect. 2004;57(4):316-320. doi:10.1016/j.jhin.2004.03.028
8. Magleby R, Westblade LF, Trzebucki A, et al. Impact Severe acute respiratory syndrome coronavirus 2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease 2019. Clin Infect Dis. 2021;73(11):e4197-e4205. doi:10.1093/cid/ciaa851
9. Buchan B, Hoff J, Gmehlin C, et al. Distribution of SARSCoV- 2 PCR cycle threshold values provide practical insight into overall and target-specific sensitivity among symptomatic patients. Am Clin Pathol. 2020;154:479-485. doi:10.1093/ajcp/aqaa133
10. He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672-675. doi:10.1038/s41591-020-0869-5
11. Zou L, Ruan F, Huang M, et al. SARS-CoV-2 Viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177-1179. doi:10.1056/NEJMc2001737
12. Singanayagam A, Patel M, Charlett A, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25(32):2001483. doi:10.2807/1560-7917.ES.2020.25.32.2001483
13. Salvatore P, Dawson P, Wadhwa A, et al. Epidemiological correlates of PCR cycles threshold values in the detection of SARS-CoV-2. Clin Infect Dis. 2021;72(11):e761-e767. doi:10.1093/cid/ciaa1469
14. Kissler S, Fauver J, Mack C, et al. Viral dynamics of SARS-CoV-2 infection and the predictive value of repeat testing. medRxiv. 2020;10.21.20217042. doi:10.1101/2020.10.21.20217042 1
5. Escobar DJ, Lanzi M, Saberi P, et al. Mitigation of a COVID-19 outbreak in a nursing home through serial testing of residents and staff. Clin Infect Dis. 2021;72(9):e394- e396. doi:10.1093/cid/ciaa1021
16. Eibner C, Krull H, Brown KM, et al. Current and projected characteristics and unique health care needs of the patient population served by the Department of Veterans Affairs. Rand Health Q. 2016;5(4):13.
17. Agha Z, Lofgren RP, VanRuiswyk JV, Layde PM. Are patients at Veterans Affairs medical centers sicker? A comparative analysis of health status and medical resource use. Arch Intern Med. 2000;160(21):3252-3257. doi:10.1001/archinte.160.21.3252
18. Morgan RO, Teal CR, Reddy SG, Ford ME, Ashton CM. Measurement in Veterans Affairs Health Services Research: veterans as a special population. Health Serv Res. 2005;40(5 Pt 2):1573-1583. doi:10.1111/j.1475-6773.2005.00448.x 1
9. Xpert Xpress SARS-CoV-2. Instructions for use. Cepheid. 302-2562, Rev. C April 2020. Accessed January 7, 2021. https://www.fda.gov/media/136314/download
20. Abbott RealTime SARS-CoV-2. Instructions for use Abbott. 09N77-95. July 2020. Accessed January 7, 2021. https:// www.fda.gov/media/136258/download
21. Petersen JM, Dalal S, Jhala D. Successful implementation of SARS-CoV-2 testing in midst of pandemic with emphasis on all phases of testing. J Clin Pathol. 2021;74:273- 278. doi:10.1136/jclinpath-2020-207175
22. United States Census Bureau. Quick Facts: Philadelphia County, Pennsylvania. Accessed April 16, 2020. https://www .census.gov/quickfacts/philadelphiacountypennsylvania
23. Centers for Disease Control and Prevention. United States COVID-19 cases, deaths, and laboratory testing (NAATS) by state, territory, and jurisdiction. Accessed April 26, 2020. https://www.cdc.gov/coronavirus/2019-ncov/cases -updates/cases-in-us.html 2
4. Petersen J, Jhala D. Ethnicity, comorbid medical conditions, and SARS-CoV-2 test cycle thresholds in the veteran population [published online ahead of print, 2021 Jul 28]. J Racial Ethn Health Disparities. 2021;1-8. doi:10.1007/s40615-021-01114-4
25. Infectious Diseases Society of America, Association for Molecular Pathology. IDSA and AMP joint statement on the use of SARS-CoV-2 PCR cycle threshold (Ct) values for clinical decision-making. Accessed August 28, 2021. https://www.idsociety.org/globalassets/idsa/public-health /covid-19/idsa-amp-statement.pdf
26. Wang J, Ng CY, Brook RH. Response to COVID-19 in Taiwan: big data analysis, new technology, and proactive testing. JAMA. 2020;323(14):1341-1342. doi:10.1001/jama.2020.3151
27. Centers for Disease Control and Prevention. Overview of testing for SARS-CoV-2, the virus that causes COVID- 19. Accessed July 28, 2021. https://www.cdc.gov /coronavirus/2019-ncov/hcp/testing-overview.html
28. Zuvekas SH, Taliaferro GS. Pathways to access: health insurance, the health care delivery system, and racial/ethnic disparities, 1996-1999. Health Aff. 2003;22(2):139-153. doi:10.1377/hlthaff.22.2.139
29. Egede LE. Race, ethnicity, culture, and disparities in health care. J Gen Intern Med. 2006;21(6):667-669. doi:10.1111/j.1525-1497.2006.0512.x
30. Institute of Medicine (US) Committee on Understanding and Eliminating Racial and Ethnic Disparities in Health Care. Smedley BD, Stith AY, Nelson AR, eds. Unequal treatment: confronting racial and ethnic disparities in health care. National Academies Press; 2003. doi:10.17226/12875
31. Ranney ML, Griffeth V, Jha AK. Critical supply shortages – the need for ventilators and personal protective equipment during the Covid-19 Pandemic. N Engl J Med. 2020;382(18):e41. doi:10.1056/NEJMp2006141
COVID-19, caused by SARS-CoV-2, is more severe in individuals with underlying illnesses. Because complete social distancing might be more difficult in nursing homes and community living centers (CLCs), public health leaders and clinicians have been concerned about the epidemiology and disease course in nursing homes even before the COVID-19 pandemic.1-7 A report of a COVID-19 outbreak in a nursing home facility in King County, Washington, documented a 33.7% overall fatality rate for residents and 52.4% among the most critically ill.4,5 The experience at King County, Washington, shows that proactive steps to identify, monitor, and apply preventive control measures is important for future outbreaks.5
Reverse transcriptase polymerase chain reaction (RT-PCR) testing produces a cycle threshold (CT) or cycle number (CN) that correlates with viral load and infectiousness. 8-14 CT/CN represents the number of RT-PCR cycles required for the fluorescent signal to cross the detection threshold (exceed background level) and is inversely proportional to the viral load. Effectively, the higher the viral load, the lower the CT/ CN value (Figure 1). Tracking CT/CN values was not documented in the Washington nursing home outbreak. Reports of COVID- 19 testing in CLCs during outbreaks are sparse, and CT/CN values and demographic distribution of these veterans has not been reported.15 The CLC veteran population, with known higher vulnerability to infection and chronic diseases, is epidemiologically different from the general nursing home population.15-18 To address these literature gaps, we present the first report of COVID- 19 testing with CT/CN value correlations in the high-risk veteran CLC population.
Methods
A retrospective review of all COVID-19 CT/CN testing at the Corporal Michael J. Crescenz Veterans Affairs Medical Center (VAMC) CLC in Philadelphia, Pennsylvania, from March 28, 2020, to April 24, 2020, was performed with a US Department of Veterans Affairs (VA) Veterans Health Information System Architecture VistA/FileMan search. Only veteran residents were included in this review. Data collected included initial and serial test results, CT/CN on positive test results, test dates, testing platform used, demographic information (age, self-reported ethnicity, and sex), and clinical follow-up information. Health records were reviewed retrospectively to identify death, the first day after diagnosis with no documented symptoms, or hospitalization status.
RT-PCR testing was performed with the Abbott RealTime SARS-CoV-2 assay on the Abbott m2000 platform and the Xpert Xpress SARS-CoV-2 assay on the Cepheid Infinity platform. The Xpert Xpress assay gave 2 CT values for the E and N2 targets on positive samples.19 For this assay to indicate a positive specimen, amplification by RT-PCR of the N2 target or both the N2 and E target is required. The Xpert Xpress assay results as presumptive positive if only the E target amplified. This assay counts a maximum of 45 cycles. The Abbott RealTime SARS-CoV-2 assay gave 1 CN derived from the RNA-dependent RNA polymerase and N targets on positive samples.20 The Abbott assay on the m2000 counts a maximum of 37 cycles. The CT/CN value is the number of cycles required by RT-PCR for the fluorescence signal to cross a threshold value exceeding background level.19,20
Samples that are negative for COVID-19 by RT-PCR do not produce a CT/CN value. Although both instruments were used for RT-PCR, the precise CT/CN values are not interchangeable and CT/CN observations over time between the 2 instruments during the disease course would be based on CT/CN value movement (general upward or downward trend) rather than absolute CT/CN differences. Both assays have been approved by emergency use authorization as qualitative tests for the presence/absence of COVID-19. Although the CT/CN value is available to laboratory staff after test completion, the CT/CN value is not reported routinely in the patient health record. All veteran patients identified on the initial review from March 28, 2020, to April 24, 2020, had all serial COVID-19 testing recorded until November 10, 2020. The CN values at the limit of detection (LOD) for the Abbott m2000 platform from the initial validation study were reviewed for reference.21
Results
Of 80 patients, 25 (31%) were COVID-19 positive over the course of testing. The study population had a mean age of 73.5 years; 92% were aged > 60 years. The group was predominantly male (79 male vs 1 female). Among the 77 patients with a stated ethnicity, 39 (51%) were African American. In comparison, 43% of residents in Philadelphia County are African American (Table).22,23 Additionally, a previously published total COVID-19 tested population by ethnicity at the same regional VAMC revealed 46.8% of tested veteran patients were African American. 24 Three patients had no stated ethnicity. Among those who tested positive, 11 were African American patients, 12 were White patients, and 2 had no stated ethnicity. Four patients tested positive on their first test. The other 21 patients were positive on repeat testing. Interestingly, 6 patients had 1 initial negative test before a positive test, 6 patients had 2, 8 patients had 3, and 1 patient had 4 initial negative tests before a positive test result. Among the 25 positive patients, 22 were either positive within 10 days of the initial negative test result or initially positive (Figure 2). Three patients who tested positive after 10 days did so at 16, 20, and 21 days after the initial negative test result. Among the 25 positive patients, 23 had initial and serial testing from both the Abbott and Xpert Xpress assays. The remaining 2 positive patients had initial and serial testing from the Abbott assay exclusively.
Only positive COVID-19 results by RTPCR produced a CT/CN value. After disease resolution with a negative test, no CT/CN value was produced with the negative test result on either testing platform. Because repeat testing after the initial positive result took place no sooner than 10 days, we observed that the CT/CN value increased after the initial positive result until the disease resolved, and a negative result was obtained (eAppendix 1, available online at doi:10.12788/fp.0276). A t test comparing the initial CT/CN value to the value more than 10 days after the initial positive showed the CT/CN was statistically significantly higher (P < .05).
Prompt repeat testing after the initial test can show a decrease in the CT/CN value because of increasing viral load before the expected increase until disease resolution if the initial test caught the infection early. Twelve patients had a negative test result between 2 serial positive results. These negative test results occurred later, near the end of the disease course. Among the 12 patients with this positive-negativepositive CT/CN pattern, 7 were symptomatic and no longer had documented symptoms or hospitalization around the time of this positive-negative-positive pattern. Four of these individuals were asymptomatic during the entire infection course. One of the 12 patients with this pattern expired with the negative result occurring on day 27 of the disease in the context of rising CT/CN. One of these 12 patients only had a presumptive positive test result on the Cepheid because it detected only the E target with a CT value of 38.7. In 1 of the 12 patients, the negative test result occurred between 2 positive test results with CT/CN values < 20 (12.05 and 19.05 for the positive tests before and after the negative result, respectively). When the initial CT/CN values was separated based on ethnicity, the average CT/CN value for African Americans (23.3) was higher than for other ethnicities (19.9), although it did not reach statistical significance (P = .35).
Ten of the 25 patients testing positive were admitted to the hospital, including 1 admitted 15 days before diagnosis (patient 20) and 1 admitted 80 days after diagnosis (patient 7). Among these 10 patients, 6 were admitted to the intensive care unit, including patient 7. None of the patients were intubated. Three of the 10 admitted patients died (patients 7, 20, and 24). Patient 7 was a 79-year-old male with a history of dementia, cerebrovascular accident, hypertension, hyperlipidemia, and chronic kidney disease with symptoms of lethargy and refusal of oral intake when he was diagnosed with COVID-19. He was admitted 80 days after diagnosis for hyponatremia and acute renal failure, with death on day 87 recorded as complications from the earlier COVID-19 infection. Patient 20, an 89-year-old male with a history of dementia, chronic kidney disease, and hyperlipidemia, had been admitted with fever, cough, and leukocytosis 17 days before COVID-19 diagnosis. He continued to be symptomatic after diagnosis with development of hypotension, dehydration, and refusal of oral intake while on comfort measures/endof- life care and died 15 days after COVID- 19 infection diagnosis. Patient 24 was a 96-year-old male with history of heart failure, hypertension, coronary artery disease, prostate carcinoma, and dementia who developed a cough at the time of diagnosis; because of his underlying condition, he remained in the CLC on comfort care. His symptoms, including hypoxia, worsened until he died 7 days after diagnosis.
Among the 25 patients, 17 were symptomatic at the time of diagnosis; the 14 initially symptomatic patients who survived improved clinically and returned to baseline. Eight of the 25 patients were asymptomatic initially and 3 developed symptoms 2 to 5 days after diagnosis. Only 1 patient who remained asymptomatic was admitted for inability to adhere to quarantine at the CLC. Review of the health records of all surviving symptomatic patients showed symptom resolution with return to baseline that corresponds to an increasing CT/CN value. A 1-tailed t test comparing the initial CT/ CN at the time of diagnosis to the last CT/CN value for symptomatic patients who recovered revealed a statistically significant increase (P < .05). For the symptomatic, symptom resolution and hospital discharge took (if required) a mean 20 days (range, 7-46). Among those who were not hospitalized, symptoms resolved in 7 to 36 days (18 days). Among those requiring hospitalization at any time (excluding patients who died or were asymptomatic), symptom and hospitalization resolution took a mean 22 days (range, 10-46). Asymptomatic patients (patients 8, 10, 15, 16, and 25) also showed increasing CT/CN value during the infection course, although there was no correlation with the continued lack of symptoms.
During the initial validation of the Abbott m2000 instrument, an LOD study included concentrations of 1000, 500, 250, 100, 70, 60, and 50 virus copies/mL (eAppendix 2, available online at doi:10.12788/fp.0276).21 The average CN at 100 virus copies/mL—the manufacturer provided LOD in the instructions for use—was 25.74.20 At a concentration of one-half that (50 virus copies/mL), the average CN was 28.39.
Discussion
This is the first study in the English literature to track CT/CN values as part of serial testing of a veteran CLC. Widescale testing and repeat screening in the absence of symptoms of nursing home residents would identify those who are infected and allow providers to track viral load clearance.9-14 CT/CN values, when serially tracked during the infection course, appear to increase with illness resolution, consistent with earlier reports that CT/CN correlates with viral load.8-14 Serial CT/CN values that are high (> 25) and continue to increase with each test suggest progression toward disease resolution or viral RNA clearance.8-14 After symptom resolution, patients can have a persistent low level of viral shedding (corresponding to a high CT/CN value).10-14,25 Near the end of disease resolution, a negative serial RT-PCR sample test before a subsequent positive might be a promising clinical sign of near disease recovery. Once the viral load is low with a CT/CN significantly higher than 25, some specimens might result as negative but turn up positive on subsequent sampling with a high CT/CN value. This pattern, with attendant high CT/CN values for the positive results, are consistent with the known effect of viral load (ie, a low viral load correlates to a high CT/CN) and adequacy of specimen collection on CT/CN values.25 If the patient’s viral load is low, the sample collected might have a viral load at or near the testing platform’s LOD.
For Abbott m2000, the manufacturer provided LOD is 100 virus copies/mL, although the instrument was able to detect virus concentrations below that level during the initial validation.20 The actual LOD of the instrument at our institution is < 100 virus copies/mL. For the Cepheid Xpert Xpress SARS-CoV-2 assay, the manufacturer-provided LOD is 250 virus copies/mL.19 An LOD study including samples below the manufacturer-provided LOD was not part of the initial validation study for the Xpert Xpress assay. Nonetheless, the virus concentration of samples with very high CT values at or near the maximum CT value of 45 is expected to be at or near the platform’s actual LOD.
If the samples collected near the end of the patient’s disease course have viral loads near these low concentrations, the encouraging positive-negative-positive pattern with high CT/CN values might be a promising sign for viral clearance. On the other hand, a positive-negative-positive pattern in the setting of low CT/CN values before and after the negative test might indicate poor sampling for the negative specimen. The back-and-forth or positive-negative-positive pattern generally appears to indicate near resolution of the infection course, although clinical correlation is necessary to rule out inadequate sampling earlier in the disease course or prolonged viral RNA shedding.9-14 In all of the surviving symptomatic patients who showed the positive-negative-positive pattern, this sign occurred around or after symptom resolution. It also is important to consider that in some patients, SARS-CoV-2 RNA might remain detectable with increasing CT/CN after symptom resolution, and samples from these patients might not result positive. Therefore, CT/CN values cannot be interpreted without considering the clinical picture.25
Studies on infectiousness and virus culture from COVID-19 samples with CT/ CN correlation have shown that patients with high CT/CN at the end of their disease course might not be as infectious.9-14,25 Because 1 patient had a presumptive positive result after the negative result, this study shows that this positive-negative-positive pattern could include presumptive positive results. Also, in the setting of a recent positive result on the same testing platform, a patient with this pattern is presumed to be positive for COVID-19 RNA because of scant viral material.
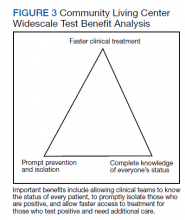
Taiwan’s public health response to the outbreak illustrates the ability to mitigate an outbreak throughout a society.26 These actions could help blunt an outbreak within a civilian nursing home population.5 Mitigation within a veteran CLC population has been documented, but the study, which focused on mitigation, did not consider CT/CN values, demographic distribution, testing access of the studied population, or laboratory findings related to disease pathophysiology.15 A key ingredient in widescale, serial testing is the availability of a rapid turnaround from testing in-house that allowed identification within 24 hours instead of several days at a reference laboratory. 15 Rapid widescale testing would allow clinical teams to optimize the Triangle of Benefit of Widescale Timely Tests for CLC (Figure 3).15 Timely laboratory testing remains pivotal for CLC veteran residents to aid successful clinical triage and management. Reporting serial CT/CN values can provide additional information to clinicians about the disease course because CT/ CN correlates with viral load, which varies based on where the patient is in the disease course.9-14 CT/CN values carry significant prognostic value, particularly with respect to intubation and mortality.8
Limitations
Important limitations to our study include the use of 2 separate RT-PCR platforms. Using different RT-PCR platforms is common in clinical laboratories trying to take advantage of the unique characteristics of different platforms—for example, turnaround time vs high throughput— to manage COVID-19 testing workflow.25 However, the exact CT/CN values obtained from each platform might not translate to the other, and the general trend (CT/CN values are rising or falling across serial tests) rather than a single value could be useful for clinical correlation. Even when the same platform is used for the serial testing, CT/CN values can be affected by adequacy of specimen collection; therefore, clinical correlation and considering the trend in CT/CN values is necessary for interpretation.10-14,25 Because of the known trend in viral dynamics, a positive specimen collected with a high CT/CN followed by a subsequent (within 2 days) positive specimen collected with a low CT/CN might be compatible with early detection of COVID- 19 infection in the appropriate clinical context. 10-14 However, detection late in the infection course or even after the symptomatic disease resolved with prolonged viral shedding might show serial positive samples with increasing CT/CN values.10-14
Patients with prolonged viral shedding might not be infectious.27 Because of the clinical correlation required for interpretation and the other factors that might affect CT/CN values, recommendations advise against using CT/CN values in clinical practice at this time, although these recommendations could change with future research.25 Serial CT/CN values have the potential, if appropriately correlated with the clinical picture, to provide useful information, such as whether the viral load of the sample is relatively high or low and increasing or decreasing.
Veterans, as a population, are more susceptible to poor health outcomes and morbidity compared with similar civilian counterparts.2,14-16 Veteran CLC patients likely would experience worse outcomes with COVID-19, including more infections, expiration, and morbidity compared with similar general population nursing homes. Similar to what had been reported for the civilian population, a trend (high CT/CN values early in the disease course with repeat testing needed to detect all positives followed by lower CT/CN value to correlate with increased viral load and then increased CT/CN value as the infection resolved) also was observed in this veteran population.
It has been extensively documented that minority groups experience decreased health care access and worse health outcomes. 28-30 Considering the critical medical supply shortages, including personal protective equipment, ventilators, and even testing supplies, there is the potential for a resource access disparity by ethnicity.28-31 Because the VA does not depend on measures of wealth and privilege such as health insurance, there was no disparity noted in access to testing by race or ethnicity at the VAMC CLC. When considering the health outcome of viral load from the measured CT/CN value, the viral loads of African American patients and those of other ethnicities was not significantly different in this study.
Conclusions
This is the first study to bring up critical points including serial CT/CN value correlation in RT-PCR tests, demographic distributions demonstrating easy and equal access in a veteran nursing home to COVID-19 testing, and clinical laboratory signs related to disease pathophysiology. Unlike other populations who have undergone serial CT/CN monitoring, nursing homes represent a particularly vulnerable population who require measures to prevent the spread and mitigate outbreaks of COVID-19.2,4,5 Test measurements obtained such as the CT/CN value during routine clinical care can provide useful information for public health, epidemiologic, or clinical purposes with appropriate correlation to clinical and other laboratory parameters. This study demonstrates early intervention of serial testing of an outbreak in a veterans nursing home with CT/CN value correlation.
COVID-19, caused by SARS-CoV-2, is more severe in individuals with underlying illnesses. Because complete social distancing might be more difficult in nursing homes and community living centers (CLCs), public health leaders and clinicians have been concerned about the epidemiology and disease course in nursing homes even before the COVID-19 pandemic.1-7 A report of a COVID-19 outbreak in a nursing home facility in King County, Washington, documented a 33.7% overall fatality rate for residents and 52.4% among the most critically ill.4,5 The experience at King County, Washington, shows that proactive steps to identify, monitor, and apply preventive control measures is important for future outbreaks.5
Reverse transcriptase polymerase chain reaction (RT-PCR) testing produces a cycle threshold (CT) or cycle number (CN) that correlates with viral load and infectiousness. 8-14 CT/CN represents the number of RT-PCR cycles required for the fluorescent signal to cross the detection threshold (exceed background level) and is inversely proportional to the viral load. Effectively, the higher the viral load, the lower the CT/ CN value (Figure 1). Tracking CT/CN values was not documented in the Washington nursing home outbreak. Reports of COVID- 19 testing in CLCs during outbreaks are sparse, and CT/CN values and demographic distribution of these veterans has not been reported.15 The CLC veteran population, with known higher vulnerability to infection and chronic diseases, is epidemiologically different from the general nursing home population.15-18 To address these literature gaps, we present the first report of COVID- 19 testing with CT/CN value correlations in the high-risk veteran CLC population.
Methods
A retrospective review of all COVID-19 CT/CN testing at the Corporal Michael J. Crescenz Veterans Affairs Medical Center (VAMC) CLC in Philadelphia, Pennsylvania, from March 28, 2020, to April 24, 2020, was performed with a US Department of Veterans Affairs (VA) Veterans Health Information System Architecture VistA/FileMan search. Only veteran residents were included in this review. Data collected included initial and serial test results, CT/CN on positive test results, test dates, testing platform used, demographic information (age, self-reported ethnicity, and sex), and clinical follow-up information. Health records were reviewed retrospectively to identify death, the first day after diagnosis with no documented symptoms, or hospitalization status.
RT-PCR testing was performed with the Abbott RealTime SARS-CoV-2 assay on the Abbott m2000 platform and the Xpert Xpress SARS-CoV-2 assay on the Cepheid Infinity platform. The Xpert Xpress assay gave 2 CT values for the E and N2 targets on positive samples.19 For this assay to indicate a positive specimen, amplification by RT-PCR of the N2 target or both the N2 and E target is required. The Xpert Xpress assay results as presumptive positive if only the E target amplified. This assay counts a maximum of 45 cycles. The Abbott RealTime SARS-CoV-2 assay gave 1 CN derived from the RNA-dependent RNA polymerase and N targets on positive samples.20 The Abbott assay on the m2000 counts a maximum of 37 cycles. The CT/CN value is the number of cycles required by RT-PCR for the fluorescence signal to cross a threshold value exceeding background level.19,20
Samples that are negative for COVID-19 by RT-PCR do not produce a CT/CN value. Although both instruments were used for RT-PCR, the precise CT/CN values are not interchangeable and CT/CN observations over time between the 2 instruments during the disease course would be based on CT/CN value movement (general upward or downward trend) rather than absolute CT/CN differences. Both assays have been approved by emergency use authorization as qualitative tests for the presence/absence of COVID-19. Although the CT/CN value is available to laboratory staff after test completion, the CT/CN value is not reported routinely in the patient health record. All veteran patients identified on the initial review from March 28, 2020, to April 24, 2020, had all serial COVID-19 testing recorded until November 10, 2020. The CN values at the limit of detection (LOD) for the Abbott m2000 platform from the initial validation study were reviewed for reference.21
Results
Of 80 patients, 25 (31%) were COVID-19 positive over the course of testing. The study population had a mean age of 73.5 years; 92% were aged > 60 years. The group was predominantly male (79 male vs 1 female). Among the 77 patients with a stated ethnicity, 39 (51%) were African American. In comparison, 43% of residents in Philadelphia County are African American (Table).22,23 Additionally, a previously published total COVID-19 tested population by ethnicity at the same regional VAMC revealed 46.8% of tested veteran patients were African American. 24 Three patients had no stated ethnicity. Among those who tested positive, 11 were African American patients, 12 were White patients, and 2 had no stated ethnicity. Four patients tested positive on their first test. The other 21 patients were positive on repeat testing. Interestingly, 6 patients had 1 initial negative test before a positive test, 6 patients had 2, 8 patients had 3, and 1 patient had 4 initial negative tests before a positive test result. Among the 25 positive patients, 22 were either positive within 10 days of the initial negative test result or initially positive (Figure 2). Three patients who tested positive after 10 days did so at 16, 20, and 21 days after the initial negative test result. Among the 25 positive patients, 23 had initial and serial testing from both the Abbott and Xpert Xpress assays. The remaining 2 positive patients had initial and serial testing from the Abbott assay exclusively.
Only positive COVID-19 results by RTPCR produced a CT/CN value. After disease resolution with a negative test, no CT/CN value was produced with the negative test result on either testing platform. Because repeat testing after the initial positive result took place no sooner than 10 days, we observed that the CT/CN value increased after the initial positive result until the disease resolved, and a negative result was obtained (eAppendix 1, available online at doi:10.12788/fp.0276). A t test comparing the initial CT/CN value to the value more than 10 days after the initial positive showed the CT/CN was statistically significantly higher (P < .05).
Prompt repeat testing after the initial test can show a decrease in the CT/CN value because of increasing viral load before the expected increase until disease resolution if the initial test caught the infection early. Twelve patients had a negative test result between 2 serial positive results. These negative test results occurred later, near the end of the disease course. Among the 12 patients with this positive-negativepositive CT/CN pattern, 7 were symptomatic and no longer had documented symptoms or hospitalization around the time of this positive-negative-positive pattern. Four of these individuals were asymptomatic during the entire infection course. One of the 12 patients with this pattern expired with the negative result occurring on day 27 of the disease in the context of rising CT/CN. One of these 12 patients only had a presumptive positive test result on the Cepheid because it detected only the E target with a CT value of 38.7. In 1 of the 12 patients, the negative test result occurred between 2 positive test results with CT/CN values < 20 (12.05 and 19.05 for the positive tests before and after the negative result, respectively). When the initial CT/CN values was separated based on ethnicity, the average CT/CN value for African Americans (23.3) was higher than for other ethnicities (19.9), although it did not reach statistical significance (P = .35).
Ten of the 25 patients testing positive were admitted to the hospital, including 1 admitted 15 days before diagnosis (patient 20) and 1 admitted 80 days after diagnosis (patient 7). Among these 10 patients, 6 were admitted to the intensive care unit, including patient 7. None of the patients were intubated. Three of the 10 admitted patients died (patients 7, 20, and 24). Patient 7 was a 79-year-old male with a history of dementia, cerebrovascular accident, hypertension, hyperlipidemia, and chronic kidney disease with symptoms of lethargy and refusal of oral intake when he was diagnosed with COVID-19. He was admitted 80 days after diagnosis for hyponatremia and acute renal failure, with death on day 87 recorded as complications from the earlier COVID-19 infection. Patient 20, an 89-year-old male with a history of dementia, chronic kidney disease, and hyperlipidemia, had been admitted with fever, cough, and leukocytosis 17 days before COVID-19 diagnosis. He continued to be symptomatic after diagnosis with development of hypotension, dehydration, and refusal of oral intake while on comfort measures/endof- life care and died 15 days after COVID- 19 infection diagnosis. Patient 24 was a 96-year-old male with history of heart failure, hypertension, coronary artery disease, prostate carcinoma, and dementia who developed a cough at the time of diagnosis; because of his underlying condition, he remained in the CLC on comfort care. His symptoms, including hypoxia, worsened until he died 7 days after diagnosis.
Among the 25 patients, 17 were symptomatic at the time of diagnosis; the 14 initially symptomatic patients who survived improved clinically and returned to baseline. Eight of the 25 patients were asymptomatic initially and 3 developed symptoms 2 to 5 days after diagnosis. Only 1 patient who remained asymptomatic was admitted for inability to adhere to quarantine at the CLC. Review of the health records of all surviving symptomatic patients showed symptom resolution with return to baseline that corresponds to an increasing CT/CN value. A 1-tailed t test comparing the initial CT/ CN at the time of diagnosis to the last CT/CN value for symptomatic patients who recovered revealed a statistically significant increase (P < .05). For the symptomatic, symptom resolution and hospital discharge took (if required) a mean 20 days (range, 7-46). Among those who were not hospitalized, symptoms resolved in 7 to 36 days (18 days). Among those requiring hospitalization at any time (excluding patients who died or were asymptomatic), symptom and hospitalization resolution took a mean 22 days (range, 10-46). Asymptomatic patients (patients 8, 10, 15, 16, and 25) also showed increasing CT/CN value during the infection course, although there was no correlation with the continued lack of symptoms.
During the initial validation of the Abbott m2000 instrument, an LOD study included concentrations of 1000, 500, 250, 100, 70, 60, and 50 virus copies/mL (eAppendix 2, available online at doi:10.12788/fp.0276).21 The average CN at 100 virus copies/mL—the manufacturer provided LOD in the instructions for use—was 25.74.20 At a concentration of one-half that (50 virus copies/mL), the average CN was 28.39.
Discussion
This is the first study in the English literature to track CT/CN values as part of serial testing of a veteran CLC. Widescale testing and repeat screening in the absence of symptoms of nursing home residents would identify those who are infected and allow providers to track viral load clearance.9-14 CT/CN values, when serially tracked during the infection course, appear to increase with illness resolution, consistent with earlier reports that CT/CN correlates with viral load.8-14 Serial CT/CN values that are high (> 25) and continue to increase with each test suggest progression toward disease resolution or viral RNA clearance.8-14 After symptom resolution, patients can have a persistent low level of viral shedding (corresponding to a high CT/CN value).10-14,25 Near the end of disease resolution, a negative serial RT-PCR sample test before a subsequent positive might be a promising clinical sign of near disease recovery. Once the viral load is low with a CT/CN significantly higher than 25, some specimens might result as negative but turn up positive on subsequent sampling with a high CT/CN value. This pattern, with attendant high CT/CN values for the positive results, are consistent with the known effect of viral load (ie, a low viral load correlates to a high CT/CN) and adequacy of specimen collection on CT/CN values.25 If the patient’s viral load is low, the sample collected might have a viral load at or near the testing platform’s LOD.
For Abbott m2000, the manufacturer provided LOD is 100 virus copies/mL, although the instrument was able to detect virus concentrations below that level during the initial validation.20 The actual LOD of the instrument at our institution is < 100 virus copies/mL. For the Cepheid Xpert Xpress SARS-CoV-2 assay, the manufacturer-provided LOD is 250 virus copies/mL.19 An LOD study including samples below the manufacturer-provided LOD was not part of the initial validation study for the Xpert Xpress assay. Nonetheless, the virus concentration of samples with very high CT values at or near the maximum CT value of 45 is expected to be at or near the platform’s actual LOD.
If the samples collected near the end of the patient’s disease course have viral loads near these low concentrations, the encouraging positive-negative-positive pattern with high CT/CN values might be a promising sign for viral clearance. On the other hand, a positive-negative-positive pattern in the setting of low CT/CN values before and after the negative test might indicate poor sampling for the negative specimen. The back-and-forth or positive-negative-positive pattern generally appears to indicate near resolution of the infection course, although clinical correlation is necessary to rule out inadequate sampling earlier in the disease course or prolonged viral RNA shedding.9-14 In all of the surviving symptomatic patients who showed the positive-negative-positive pattern, this sign occurred around or after symptom resolution. It also is important to consider that in some patients, SARS-CoV-2 RNA might remain detectable with increasing CT/CN after symptom resolution, and samples from these patients might not result positive. Therefore, CT/CN values cannot be interpreted without considering the clinical picture.25
Studies on infectiousness and virus culture from COVID-19 samples with CT/ CN correlation have shown that patients with high CT/CN at the end of their disease course might not be as infectious.9-14,25 Because 1 patient had a presumptive positive result after the negative result, this study shows that this positive-negative-positive pattern could include presumptive positive results. Also, in the setting of a recent positive result on the same testing platform, a patient with this pattern is presumed to be positive for COVID-19 RNA because of scant viral material.
Taiwan’s public health response to the outbreak illustrates the ability to mitigate an outbreak throughout a society.26 These actions could help blunt an outbreak within a civilian nursing home population.5 Mitigation within a veteran CLC population has been documented, but the study, which focused on mitigation, did not consider CT/CN values, demographic distribution, testing access of the studied population, or laboratory findings related to disease pathophysiology.15 A key ingredient in widescale, serial testing is the availability of a rapid turnaround from testing in-house that allowed identification within 24 hours instead of several days at a reference laboratory. 15 Rapid widescale testing would allow clinical teams to optimize the Triangle of Benefit of Widescale Timely Tests for CLC (Figure 3).15 Timely laboratory testing remains pivotal for CLC veteran residents to aid successful clinical triage and management. Reporting serial CT/CN values can provide additional information to clinicians about the disease course because CT/ CN correlates with viral load, which varies based on where the patient is in the disease course.9-14 CT/CN values carry significant prognostic value, particularly with respect to intubation and mortality.8
Limitations
Important limitations to our study include the use of 2 separate RT-PCR platforms. Using different RT-PCR platforms is common in clinical laboratories trying to take advantage of the unique characteristics of different platforms—for example, turnaround time vs high throughput— to manage COVID-19 testing workflow.25 However, the exact CT/CN values obtained from each platform might not translate to the other, and the general trend (CT/CN values are rising or falling across serial tests) rather than a single value could be useful for clinical correlation. Even when the same platform is used for the serial testing, CT/CN values can be affected by adequacy of specimen collection; therefore, clinical correlation and considering the trend in CT/CN values is necessary for interpretation.10-14,25 Because of the known trend in viral dynamics, a positive specimen collected with a high CT/CN followed by a subsequent (within 2 days) positive specimen collected with a low CT/CN might be compatible with early detection of COVID- 19 infection in the appropriate clinical context. 10-14 However, detection late in the infection course or even after the symptomatic disease resolved with prolonged viral shedding might show serial positive samples with increasing CT/CN values.10-14
Patients with prolonged viral shedding might not be infectious.27 Because of the clinical correlation required for interpretation and the other factors that might affect CT/CN values, recommendations advise against using CT/CN values in clinical practice at this time, although these recommendations could change with future research.25 Serial CT/CN values have the potential, if appropriately correlated with the clinical picture, to provide useful information, such as whether the viral load of the sample is relatively high or low and increasing or decreasing.
Veterans, as a population, are more susceptible to poor health outcomes and morbidity compared with similar civilian counterparts.2,14-16 Veteran CLC patients likely would experience worse outcomes with COVID-19, including more infections, expiration, and morbidity compared with similar general population nursing homes. Similar to what had been reported for the civilian population, a trend (high CT/CN values early in the disease course with repeat testing needed to detect all positives followed by lower CT/CN value to correlate with increased viral load and then increased CT/CN value as the infection resolved) also was observed in this veteran population.
It has been extensively documented that minority groups experience decreased health care access and worse health outcomes. 28-30 Considering the critical medical supply shortages, including personal protective equipment, ventilators, and even testing supplies, there is the potential for a resource access disparity by ethnicity.28-31 Because the VA does not depend on measures of wealth and privilege such as health insurance, there was no disparity noted in access to testing by race or ethnicity at the VAMC CLC. When considering the health outcome of viral load from the measured CT/CN value, the viral loads of African American patients and those of other ethnicities was not significantly different in this study.
Conclusions
This is the first study to bring up critical points including serial CT/CN value correlation in RT-PCR tests, demographic distributions demonstrating easy and equal access in a veteran nursing home to COVID-19 testing, and clinical laboratory signs related to disease pathophysiology. Unlike other populations who have undergone serial CT/CN monitoring, nursing homes represent a particularly vulnerable population who require measures to prevent the spread and mitigate outbreaks of COVID-19.2,4,5 Test measurements obtained such as the CT/CN value during routine clinical care can provide useful information for public health, epidemiologic, or clinical purposes with appropriate correlation to clinical and other laboratory parameters. This study demonstrates early intervention of serial testing of an outbreak in a veterans nursing home with CT/CN value correlation.
1. Chen T, Wu D, Chen H, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. doi:10.1136/bmj.m1091
2. Tsan L, Davis C, Langberg R, et al. Prevalence of nursing home-associated infections in the Department of Veterans Affairs nursing home care units. Am J Infect Control. 2008;36(3):173-179. doi:10.1016/j.ajic.2007.06.008
3. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054-1062. doi:10.1016/S0140-6736(20)30566-3
4. Arentz M, Yim E, Klaff L, et al. Characteristics and outcomes of 21 critically ill patients with COVID-19 in Washington State. JAMA. 2020;323(16):1612-1614. doi:10.1001/jama.2020.4326
5. McMichael TM, Currie DW, Clark S, et al. Public Health–Seattle and King County, EvergreenHealth, and CDC COVID-19 Investigation Team. Epidemiology of Covid-19 in a long-term care facility in King County, Washington. N Engl J Med. 2020;382(21):2005-2011. doi:10.1056/NEJMoa2005412
6. Childs A, Zullo AR, Joyce NR, et al. The burden of respiratory infections among older adults in long-term care: a systematic review. BMC Geriatr. 2019;19(1):210. doi:10.1186/s12877-019-1236-6
7. Eriksen HM, Iversen BG, Aavitsland PJ. Prevalence of nosocomial infections and use of antibiotics in long-term care facilities in Norway, 2002 and 2003. Hosp Infect. 2004;57(4):316-320. doi:10.1016/j.jhin.2004.03.028
8. Magleby R, Westblade LF, Trzebucki A, et al. Impact Severe acute respiratory syndrome coronavirus 2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease 2019. Clin Infect Dis. 2021;73(11):e4197-e4205. doi:10.1093/cid/ciaa851
9. Buchan B, Hoff J, Gmehlin C, et al. Distribution of SARSCoV- 2 PCR cycle threshold values provide practical insight into overall and target-specific sensitivity among symptomatic patients. Am Clin Pathol. 2020;154:479-485. doi:10.1093/ajcp/aqaa133
10. He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672-675. doi:10.1038/s41591-020-0869-5
11. Zou L, Ruan F, Huang M, et al. SARS-CoV-2 Viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177-1179. doi:10.1056/NEJMc2001737
12. Singanayagam A, Patel M, Charlett A, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25(32):2001483. doi:10.2807/1560-7917.ES.2020.25.32.2001483
13. Salvatore P, Dawson P, Wadhwa A, et al. Epidemiological correlates of PCR cycles threshold values in the detection of SARS-CoV-2. Clin Infect Dis. 2021;72(11):e761-e767. doi:10.1093/cid/ciaa1469
14. Kissler S, Fauver J, Mack C, et al. Viral dynamics of SARS-CoV-2 infection and the predictive value of repeat testing. medRxiv. 2020;10.21.20217042. doi:10.1101/2020.10.21.20217042 1
5. Escobar DJ, Lanzi M, Saberi P, et al. Mitigation of a COVID-19 outbreak in a nursing home through serial testing of residents and staff. Clin Infect Dis. 2021;72(9):e394- e396. doi:10.1093/cid/ciaa1021
16. Eibner C, Krull H, Brown KM, et al. Current and projected characteristics and unique health care needs of the patient population served by the Department of Veterans Affairs. Rand Health Q. 2016;5(4):13.
17. Agha Z, Lofgren RP, VanRuiswyk JV, Layde PM. Are patients at Veterans Affairs medical centers sicker? A comparative analysis of health status and medical resource use. Arch Intern Med. 2000;160(21):3252-3257. doi:10.1001/archinte.160.21.3252
18. Morgan RO, Teal CR, Reddy SG, Ford ME, Ashton CM. Measurement in Veterans Affairs Health Services Research: veterans as a special population. Health Serv Res. 2005;40(5 Pt 2):1573-1583. doi:10.1111/j.1475-6773.2005.00448.x 1
9. Xpert Xpress SARS-CoV-2. Instructions for use. Cepheid. 302-2562, Rev. C April 2020. Accessed January 7, 2021. https://www.fda.gov/media/136314/download
20. Abbott RealTime SARS-CoV-2. Instructions for use Abbott. 09N77-95. July 2020. Accessed January 7, 2021. https:// www.fda.gov/media/136258/download
21. Petersen JM, Dalal S, Jhala D. Successful implementation of SARS-CoV-2 testing in midst of pandemic with emphasis on all phases of testing. J Clin Pathol. 2021;74:273- 278. doi:10.1136/jclinpath-2020-207175
22. United States Census Bureau. Quick Facts: Philadelphia County, Pennsylvania. Accessed April 16, 2020. https://www .census.gov/quickfacts/philadelphiacountypennsylvania
23. Centers for Disease Control and Prevention. United States COVID-19 cases, deaths, and laboratory testing (NAATS) by state, territory, and jurisdiction. Accessed April 26, 2020. https://www.cdc.gov/coronavirus/2019-ncov/cases -updates/cases-in-us.html 2
4. Petersen J, Jhala D. Ethnicity, comorbid medical conditions, and SARS-CoV-2 test cycle thresholds in the veteran population [published online ahead of print, 2021 Jul 28]. J Racial Ethn Health Disparities. 2021;1-8. doi:10.1007/s40615-021-01114-4
25. Infectious Diseases Society of America, Association for Molecular Pathology. IDSA and AMP joint statement on the use of SARS-CoV-2 PCR cycle threshold (Ct) values for clinical decision-making. Accessed August 28, 2021. https://www.idsociety.org/globalassets/idsa/public-health /covid-19/idsa-amp-statement.pdf
26. Wang J, Ng CY, Brook RH. Response to COVID-19 in Taiwan: big data analysis, new technology, and proactive testing. JAMA. 2020;323(14):1341-1342. doi:10.1001/jama.2020.3151
27. Centers for Disease Control and Prevention. Overview of testing for SARS-CoV-2, the virus that causes COVID- 19. Accessed July 28, 2021. https://www.cdc.gov /coronavirus/2019-ncov/hcp/testing-overview.html
28. Zuvekas SH, Taliaferro GS. Pathways to access: health insurance, the health care delivery system, and racial/ethnic disparities, 1996-1999. Health Aff. 2003;22(2):139-153. doi:10.1377/hlthaff.22.2.139
29. Egede LE. Race, ethnicity, culture, and disparities in health care. J Gen Intern Med. 2006;21(6):667-669. doi:10.1111/j.1525-1497.2006.0512.x
30. Institute of Medicine (US) Committee on Understanding and Eliminating Racial and Ethnic Disparities in Health Care. Smedley BD, Stith AY, Nelson AR, eds. Unequal treatment: confronting racial and ethnic disparities in health care. National Academies Press; 2003. doi:10.17226/12875
31. Ranney ML, Griffeth V, Jha AK. Critical supply shortages – the need for ventilators and personal protective equipment during the Covid-19 Pandemic. N Engl J Med. 2020;382(18):e41. doi:10.1056/NEJMp2006141
1. Chen T, Wu D, Chen H, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. doi:10.1136/bmj.m1091
2. Tsan L, Davis C, Langberg R, et al. Prevalence of nursing home-associated infections in the Department of Veterans Affairs nursing home care units. Am J Infect Control. 2008;36(3):173-179. doi:10.1016/j.ajic.2007.06.008
3. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054-1062. doi:10.1016/S0140-6736(20)30566-3
4. Arentz M, Yim E, Klaff L, et al. Characteristics and outcomes of 21 critically ill patients with COVID-19 in Washington State. JAMA. 2020;323(16):1612-1614. doi:10.1001/jama.2020.4326
5. McMichael TM, Currie DW, Clark S, et al. Public Health–Seattle and King County, EvergreenHealth, and CDC COVID-19 Investigation Team. Epidemiology of Covid-19 in a long-term care facility in King County, Washington. N Engl J Med. 2020;382(21):2005-2011. doi:10.1056/NEJMoa2005412
6. Childs A, Zullo AR, Joyce NR, et al. The burden of respiratory infections among older adults in long-term care: a systematic review. BMC Geriatr. 2019;19(1):210. doi:10.1186/s12877-019-1236-6
7. Eriksen HM, Iversen BG, Aavitsland PJ. Prevalence of nosocomial infections and use of antibiotics in long-term care facilities in Norway, 2002 and 2003. Hosp Infect. 2004;57(4):316-320. doi:10.1016/j.jhin.2004.03.028
8. Magleby R, Westblade LF, Trzebucki A, et al. Impact Severe acute respiratory syndrome coronavirus 2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease 2019. Clin Infect Dis. 2021;73(11):e4197-e4205. doi:10.1093/cid/ciaa851
9. Buchan B, Hoff J, Gmehlin C, et al. Distribution of SARSCoV- 2 PCR cycle threshold values provide practical insight into overall and target-specific sensitivity among symptomatic patients. Am Clin Pathol. 2020;154:479-485. doi:10.1093/ajcp/aqaa133
10. He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672-675. doi:10.1038/s41591-020-0869-5
11. Zou L, Ruan F, Huang M, et al. SARS-CoV-2 Viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177-1179. doi:10.1056/NEJMc2001737
12. Singanayagam A, Patel M, Charlett A, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25(32):2001483. doi:10.2807/1560-7917.ES.2020.25.32.2001483
13. Salvatore P, Dawson P, Wadhwa A, et al. Epidemiological correlates of PCR cycles threshold values in the detection of SARS-CoV-2. Clin Infect Dis. 2021;72(11):e761-e767. doi:10.1093/cid/ciaa1469
14. Kissler S, Fauver J, Mack C, et al. Viral dynamics of SARS-CoV-2 infection and the predictive value of repeat testing. medRxiv. 2020;10.21.20217042. doi:10.1101/2020.10.21.20217042 1
5. Escobar DJ, Lanzi M, Saberi P, et al. Mitigation of a COVID-19 outbreak in a nursing home through serial testing of residents and staff. Clin Infect Dis. 2021;72(9):e394- e396. doi:10.1093/cid/ciaa1021
16. Eibner C, Krull H, Brown KM, et al. Current and projected characteristics and unique health care needs of the patient population served by the Department of Veterans Affairs. Rand Health Q. 2016;5(4):13.
17. Agha Z, Lofgren RP, VanRuiswyk JV, Layde PM. Are patients at Veterans Affairs medical centers sicker? A comparative analysis of health status and medical resource use. Arch Intern Med. 2000;160(21):3252-3257. doi:10.1001/archinte.160.21.3252
18. Morgan RO, Teal CR, Reddy SG, Ford ME, Ashton CM. Measurement in Veterans Affairs Health Services Research: veterans as a special population. Health Serv Res. 2005;40(5 Pt 2):1573-1583. doi:10.1111/j.1475-6773.2005.00448.x 1
9. Xpert Xpress SARS-CoV-2. Instructions for use. Cepheid. 302-2562, Rev. C April 2020. Accessed January 7, 2021. https://www.fda.gov/media/136314/download
20. Abbott RealTime SARS-CoV-2. Instructions for use Abbott. 09N77-95. July 2020. Accessed January 7, 2021. https:// www.fda.gov/media/136258/download
21. Petersen JM, Dalal S, Jhala D. Successful implementation of SARS-CoV-2 testing in midst of pandemic with emphasis on all phases of testing. J Clin Pathol. 2021;74:273- 278. doi:10.1136/jclinpath-2020-207175
22. United States Census Bureau. Quick Facts: Philadelphia County, Pennsylvania. Accessed April 16, 2020. https://www .census.gov/quickfacts/philadelphiacountypennsylvania
23. Centers for Disease Control and Prevention. United States COVID-19 cases, deaths, and laboratory testing (NAATS) by state, territory, and jurisdiction. Accessed April 26, 2020. https://www.cdc.gov/coronavirus/2019-ncov/cases -updates/cases-in-us.html 2
4. Petersen J, Jhala D. Ethnicity, comorbid medical conditions, and SARS-CoV-2 test cycle thresholds in the veteran population [published online ahead of print, 2021 Jul 28]. J Racial Ethn Health Disparities. 2021;1-8. doi:10.1007/s40615-021-01114-4
25. Infectious Diseases Society of America, Association for Molecular Pathology. IDSA and AMP joint statement on the use of SARS-CoV-2 PCR cycle threshold (Ct) values for clinical decision-making. Accessed August 28, 2021. https://www.idsociety.org/globalassets/idsa/public-health /covid-19/idsa-amp-statement.pdf
26. Wang J, Ng CY, Brook RH. Response to COVID-19 in Taiwan: big data analysis, new technology, and proactive testing. JAMA. 2020;323(14):1341-1342. doi:10.1001/jama.2020.3151
27. Centers for Disease Control and Prevention. Overview of testing for SARS-CoV-2, the virus that causes COVID- 19. Accessed July 28, 2021. https://www.cdc.gov /coronavirus/2019-ncov/hcp/testing-overview.html
28. Zuvekas SH, Taliaferro GS. Pathways to access: health insurance, the health care delivery system, and racial/ethnic disparities, 1996-1999. Health Aff. 2003;22(2):139-153. doi:10.1377/hlthaff.22.2.139
29. Egede LE. Race, ethnicity, culture, and disparities in health care. J Gen Intern Med. 2006;21(6):667-669. doi:10.1111/j.1525-1497.2006.0512.x
30. Institute of Medicine (US) Committee on Understanding and Eliminating Racial and Ethnic Disparities in Health Care. Smedley BD, Stith AY, Nelson AR, eds. Unequal treatment: confronting racial and ethnic disparities in health care. National Academies Press; 2003. doi:10.17226/12875
31. Ranney ML, Griffeth V, Jha AK. Critical supply shortages – the need for ventilators and personal protective equipment during the Covid-19 Pandemic. N Engl J Med. 2020;382(18):e41. doi:10.1056/NEJMp2006141
Reducing False-Positive Results With Fourth-Generation HIV Testing at a Veterans Affairs Medical Center
Ever since the first clinical reports of patients with AIDS in 1981, there have been improvements both in the knowledge base of the pathogenesis of HIV in causing AIDS as well as a progressive refinement in the test methodologies used to diagnose this illness.1-3 Given that there are both public health and clinical benefits in earlier diagnosis and treatment of patients with available antiretroviral therapies, universal screening with opt-out consent has been a standard of practice recommendation by the Centers of Disease Control and Prevention (CDC) since 2006; universal screening with opt-out consent also has been recommended by the US Preventative Task Force and has been widely implemented.4-7
HIV Screening
While HIV screening assays have evolved to be accurate with very high sensitivities and specificities, false-positive results are a significant issue both currently and historically.8-16 The use of an HIV assay on a low prevalence population predictably reduces the positive predictive value (PPV) of even an otherwise accurate assay.8-23 In light of this, laboratory HIV testing algorithms include confirmatory testing to increase the likelihood that the correct diagnosis is being rendered.
The fourth-generation assay has been shown to be more sensitive and specific compared with that of the third-generation assay due to the addition of detection of p24 antigen and the refinement of the antigenic targets for the antibody detection.6,8,11-13,18-20,22 Due to these improvements, in the general population, increased sensitivity/specificity with a reduction in both false positives and false negatives have been reported.
It has been observed in the nonveteran population that switching from the older third-generation to a more sensitive and specific fourth-generation HIV screening assay has reduced the false-positive screening rate.18,19,22 For instance, Muthukumar and colleagues demonstrated a false-positive rate of only 2 out of 99 (2%) tested specimens for the fourth-generation ARCHITECT HIV Ag/Ab Combo assay vs 9 out of 99 tested specimens (9%) for the third-generation ADVIA Centaur HIV 1/O/2 Enhanced assay.18 In addition, it has been noted that fourth-generation HIV screening assays can reduce the window period by detecting HIV infection sooner after initial acute infection.19 Mitchell and colleagues demonstrated even highly specific fourth-generation HIV assays with specificities estimated at 99.7% can have PPVs as low as 25.0% if used in a population of low HIV prevalence (such as a 0.1% prevalence population).19 However, the veteran population has been documented to differ significantly on a number of population variables, including severity of disease and susceptibility to infections, and as a result extrapolation of these data from the general population may be limited.24-26 To our knowledge, this article represents the first study directly examining the reduction in false-positive results with the switch to a fourth-generation HIV generation assay from a third-generation assay for the veteran patient population at a regional US Department of Veterans Affairs (VA) medical center (VAMC).8,11
Methods
Quality assurance documents on test volume were retrospectively reviewed to obtain the number of HIV screening tests that were performed by the laboratory at the Corporal Michael J. Crescenz VAMC (CMJCVAMC) in Philadelphia, Pennsylvania, between March 1, 2016 and February 28, 2017, prior to implementation of the fourth-generation assay. The study also include results from the first year of use of the fourth-generation assay (March 1, 2017 to February 28, 2018). In addition, paper quality assurance records of all positive screening results during those periods were reviewed and manually counted for the abstract presentation of these data.
For assurance of accuracy, a search of all HIV testing assays using Veterans Health Information Systems and Technology Architecture and FileMan also was performed, and the results were compared to records in the Computerized Patient Record System (CPRS). Any discrepancies in the numbers of test results generated by both searches were investigated, and data for the manuscript were derived from records associating tests with particular patients. Only results from patient samples were considered for the electronic search. Quality samples that did not correspond to a true patient as identified in CPRS or same time patient sample duplicates were excluded from the calculations. Basic demographic data (age, ethnicity, and gender) were obtained from this FileMan search. The third-generation assay was the Ortho-Clinical Diagnostics Vitros, and the fourth-generation assay was the Abbott Architect.
To interpret the true HIV result of each sample with a reactive or positive screening result, the CDC laboratory HIV testing algorithm was followed and reviewed with a clinical pathologist or microbiologist director.12,13 All specimens interpreted as HIV positive by the pathologist or microbiologist director were discussed with the clinical health care provider at the time of the test with results added to CPRS after all testing was complete and discussions had taken place. All initially reactive specimens (confirmed with retesting in duplicate on the screening platform with at least 1 repeat reactive result) were further tested with the Bio-Rad Geenius HIV 1/2 Supplemental Assay, which screens for both HIV-1 and HIV-2 antibodies. Specimens with reactive results by this supplemental assay were interpreted as positive for HIV based on the CDC laboratory HIV testing algorithm. Specimens with negative or indeterminant results by the supplemental assay then underwent HIV-1 nucleic acid testing (NAT) using the Roche Diagnostics COBAS AmpliPrep/COBAS TaqMan HIV-1 Test v2.0. Specimens with viral load detected on NAT were positive for HIV infection, while specimens with viral load not detected on NAT testing were interpreted as negative for HIV-1 infection. Although there were no HIV-2 positive or indeterminant specimens during the study period, HIV-2 reactivity also would have been interpreted per the CDC laboratory HIV testing algorithm. Specimens with inadequate volume to complete all testing steps would be interpreted as indeterminant for HIV with request for additional specimen to complete testing. All testing platforms used for HIV testing in the laboratory had been properly validated prior to use.
The number of false positives and indeterminant results was tabulated in Microsoft Excel by month throughout the study period alongside the total number of HIV screening tests performed. Statistical analyses to verify statistical significance was performed by 1-tailed homoscedastic t test calculation using Excel.
Results
From March 1, 2016 to February 28, 2017, 7,516 specimens were screened for HIV, using the third-generation assay, and 52 specimens tested positive for HIV. On further review of these reactive specimens per the CDC laboratory testing algorithm, 24 tests were true positive and 28 were false positives with a PPV of 46% (24/52) (Figure 1).
From March 1, 2017 to February 28, 2018, 7,802 specimens were screened for HIV using a fourth-generation assay and 23 tested positive for HIV. On further review of these reactive specimens per the CDC laboratory testing algorithm, 16 were true positive and 7 were false positives with a PPV of 70% (16/23).
The fourth-generation assay was more specific when compared with the third-generation assay (0.09% vs 0.37%, respectively) with a 75.7% decrease in the false-positivity rate after the implementation of fourth-generation testing. The decreased number of false-positive test results per month with the fourth-generation test implementation was statistically significant (P = .002). The mean (SD) number of false-positive test results for the third-generation assay was 2.3 (1.7) per month, while the fourth-generation assay only had a mean (SD) of 0.58 (0.9) false positives monthly. The decrease in the percentage of false positives per month with the implementation of the fourth-generation assay also was statistically significant (P = .002) (Figure 2).
For population-based reference of the tested population at CMJCVAMC, there was a FileMan search for basic demographic data of patients for the HIV specimens screened by the third- or fourth-generation test (Table). For the population tested by the third-generation assay, 1,114 out of the 7,516 total tested population did not have readily available demographic information by the FileMan search as the specimens originated outside of the facility. For 6,402 of 7,516 patients tested by the third-generation assay with demographic information, the age ranged from 25 to 97 years with a mean of 57 years. This population of 6,402 was 88% male (n = 5,639), 50% African American (n = 3,220) and 43% White (n = 2,756). For the population tested by the fourth-generation assay, 993 of 7,802 total tested population did not have readily available demographic information by the FileMan search as the specimens originated outside of the facility. For the 6,809 of 7,802 patients tested by the fourth-generation assay with demographic information, the age ranged from 24 to 97 years with a mean age of 56 years. This population was 88% male (n = 5,971), 47% African American (n = 3,189), and 46% White (n = 3,149).
Discussion
Current practice guidelines from the CDC and the US Preventive Services Task Force recommend universal screening of the population for HIV infection.5,6 As the general population to be screened would normally have a low prevalence of HIV infection, the risk of a false positive on the initial screen is significant.17 Indeed, the CMJCVAMC experience has been that with the third-generation screening assay, the number of false-positive test results outnumbered the number of true-positive test results. Even with the fourth-generation assay, approximately one-third of the results were false positives. These results are similar to those observed in studies involving nonveteran populations in which the implementation of a fourth-generation screening assay led to significantly fewer false-positive results.18
For laboratories that do not follows CDC testing algorithm guidelines, each false-positive screening result represents a potential opportunity for a HIV misdiagnosis.Even in laboratories with proper procedures in place, false-positive results have consequences for the patients and for the cost-effectiveness of laboratory operations.9-11,18 As per CDC HIV testing guidelines, all positive screening results should be retested, which leads to additional use of technologist time and reagents. After this additional testing is performed and reviewed appropriately, only then can an appropriate final laboratory diagnosis be rendered that meets the standard of laboratory care.
Cost Savings
As observed at CMJCVAMC, the use of a fourth-generation assay with increased sensitivity/specificity led to a reduction in these false-positive results, which improved laboratory efficiency and avoided wasted resources for confirmatory tests.11,18 Cost savings at CMJCVAMC from the implementation of the fourth-generation assay would include technologist time and reagent cost. Generalizable technologist time costs at any institution would include the time needed to perform the confirmatory HIV-1/HIV-2 antibody differentiation assay (slightly less than 1 hour at CMJCVAMC per specimen) and the time needed to perform the viral load assay (about 6 hours to run a batch of 24 tests at CMJCVAMC). We calculated that confirmatory testing cost $184.51 per test at CMJCVAMC. Replacing the third-generation assay with the more sensitive and specific fourth-generation test saved an estimated $3,875 annually. This cost savings does not even consider savings in the pathologist/director’s time for reviewing HIV results after the completion of the algorithm or the clinician/patient costs or anxiety while waiting for results of the confirmatory sequence of tests.
As diagnosis of HIV can have a significant psychological impact on the patient, it is important to ensure the diagnosis conveyed is correct.27 The provision of an HIV diagnosis to a patient has been described as a traumatic stressor capable of causing psychological harm; this harm should ideally be avoided if the HIV diagnosis is not accurate. There can be a temptation, when presented with a positive or reactive screening test that is known to come from an instrument or assay with a very high sensitivity and specificity, to present this result as a diagnosis to the patient. However, a false diagnosis from a false-positive screen would not only be harmful, but given the low prevalence of the disease in the screened population, would happen fairly frequently; in some settings the number of false positives may actually outnumber the number of true positive test results.
Better screening assays with greater specificity (even fractions of a percentage, given that specificities are already > 99%) would help reduce the number of false positives and reduce the number of potential enticements to convey an incorrect diagnosis. Therefore, by adding an additional layer of safety through greater specificity, the fourth-generation assay implementation helped improve the diagnostic safety of the laboratory and reduced the significant error risk to the clinician who would ultimately bear responsibility for conveying the HIV diagnoses to the patient. Given the increased prevalence of psychological and physical ailments in veterans, it may be even more important to ensure the diagnosis is correct to avoid increased psychological harm.27,28
Veteran Population
For the general population, the fourth-generation assay has been shown to be more sensitive and specific when compared with the third-generation assay due to the addition of detection of p24 antigen and the refinement of the antigenic targets for the antibody detection.6,8,11-13,18-20,22 However, the veteran population that receives VA medical care differs significantly from the nonveteran general population. Compared with nonveterans, veterans tend to have generally poorer health status, more comorbid conditions, and greater need to use medical resources.24-26 In addition, veterans also may differ in sociodemographic status, race, ethnicity, and gender.24-26
VA research in the veteran population is unique, and veterans who use VA health care services are an even more highly selected subpopulation.26 Conclusions made from studies of the general population may not always be applicable to the veteran population treated by VA health care services due to these population differences. Therefore, specific studies tailored to this special veteran population in the specific VA health care setting are essential to ensure that the results of the general population truly and definitively apply to the veteran population.
While the false-positive risk is most closely associated with testing in a population of low prevalence, it also should be noted that false-positive screening results also can occur in high-risk individuals, such as an individual on preexposure prophylaxis (PrEP) for continuous behavior that places the individual at high risk of HIV acquisition.8,29 The false-positive result in these cases can lead to a conundrum for the clinician, and the differential diagnosis should consider both detection of very early infection as well as false positive. Interventions could include either stopping PrEP and treating for presumed early primary infection with HIV or continuing the PrEP. These interventions all have the potential to impact the patient whether through the production of resistant HIV virus due to the inadvertent provision of an inadequate treatment regimen, increased risk of infection if taken off PrEP as the patient may likely continue the behavior regardless, or the risks carried by the administration of additional antiretroviral therapies for the complete empiric therapy. Cases of an individual on PrEP who had a false-positive HIV screening test has been reported previously both within and outside the veteran population.8 Better screening tests with greater sensitivity/specificity can only help in guiding better patient care.
Limitations
This quality assurance study was limited to retrospectively identifying the improvement in the false-positive rate on the transition from the third-generation to the more advanced fourth-generation HIV screen. False-positive screen cases could be easily picked up on review of the confirmatory testing per the CDC laboratory HIV testing algorithm.12,13 This study also was a retrospective review of clinically ordered and indicated testing; as a result, without confirmatory testing performed on all negative screen cases, a false-negative rate would not be calculable.
This study also was restricted to only the population being treated in a VA health care setting. This population is known to be different from the general population.24-26
Conclusions
The switch to a fourth-generation assay resulted in a significant reduction in false-positive test results for veteran patients at CMJCVAMC. This reduction in false-positive screening not only reduced laboratory workload due to the necessary confirmatory testing and subsequent review, but also saved costs for technologist’s time and reagents. While this reduction in false-positive results has been documented in nonveteran populations, this is the first study specifically on a veteran population treated at a VAMC.8,11,18 This study confirms previously documented findings of improvement in the false-positive rate of HIV screening tests with the change from third-generation to fourth-generation assay for a veteran population.24
1. Feinberg MB. Changing the natural history of HIV disease. Lancet. 1996;348(9022):239-246. doi:10.1016/s0140-6736(96)06231-9.
2. Alexander TS. Human immunodeficiency virus diagnostic testing: 30 years of evolution. Clin Vaccine Immunol. 2016;23(4):249-253. Published 2016 Apr 4. doi:10.1128/CVI.00053-16
3. Mortimer PP, Parry JV, Mortimer JY. Which anti-HTLV III/LAV assays for screening and confirmatory testing?. Lancet. 1985;2(8460):873-877. doi:10.1016/s0140-6736(85)90136-9
4. Holmberg SD, Palella FJ Jr, Lichtenstein KA, Havlir DV. The case for earlier treatment of HIV infection [published correction appears in Clin Infect Dis. 2004 Dec 15;39(12):1869]. Clin Infect Dis. 2004;39(11):1699-1704. doi:10.1086/425743
5. US Preventive Services Task Force, Owens DK, Davidson KW, et al. Screening for HIV Infection: US Preventive Services Task Force Recommendation Statement. JAMA. 2019;321(23):2326-2336. doi:10.1001/jama.2019.6587
6. Branson BM, Handsfield HH, Lampe MA, et al. Revised recommendations for HIV testing of adults, adolescents, and pregnant women in health-care settings. MMWR Recomm Rep. 2006;55(RR-14):1-CE4.
7. Bayer R, Philbin M, Remien RH. The end of written informed consent for HIV testing: not with a bang but a whimper. Am J Public Health. 2017;107(8):1259-1265. doi:10.2105/AJPH.2017.303819
8. Petersen J, Jhala D. Its not HIV! The pitfall of unconfirmed positive HIV screening assays. Abstract presented at: Annual Meeting Pennsylvania Association of Pathologists; April 14, 2018.
9. Wood RW, Dunphy C, Okita K, Swenson P. Two “HIV-infected” persons not really infected. Arch Intern Med. 2003;163(15):1857-1859. doi:10.1001/archinte.163.15.1857
10. Permpalung N, Ungprasert P, Chongnarungsin D, Okoli A, Hyman CL. A diagnostic blind spot: acute infectious mononucleosis or acute retroviral syndrome. Am J Med. 2013;126(9):e5-e6. doi:10.1016/j.amjmed.2013.03.017
11. Dalal S, Petersen J, Luta D, Jhala D. Third- to fourth-generation HIV testing: reduction in false-positive results with the new way of testing, the Corporal Michael J. Crescenz Veteran Affairs Medical Center (CMCVAMC) Experience. Am J Clin Pathol.2018;150(suppl 1):S70-S71. doi:10.1093/ajcp/aqy093.172
12. Centers for Disease Control and Prevention. Laboratory testing for the diagnosis of HIV infection: updated recommendations. Published June 27, 2014. Accessed April 14, 2021. doi:10.15620/cdc.23447
13. Centers for Disease Control and Prevention. 2018 quick reference guide: recommended laboratory HIV testing algorithm for serum or plasma specimens. Updated January 2018. Accessed April 14, 202. https://stacks.cdc.gov/view/cdc/50872
14. Masciotra S, McDougal JS, Feldman J, Sprinkle P, Wesolowski L, Owen SM. Evaluation of an alternative HIV diagnostic algorithm using specimens from seroconversion panels and persons with established HIV infections. J Clin Virol. 2011;52(suppl 1):S17-S22. doi:10.1016/j.jcv.2011.09.011
15. Morton A. When lab tests lie … heterophile antibodies. Aust Fam Physician. 2014;43(6):391-393.
16. Spencer DV, Nolte FS, Zhu Y. Heterophilic antibody interference causing false-positive rapid human immunodeficiency virus antibody testing. Clin Chim Acta. 2009;399(1-2):121-122. doi:10.1016/j.cca.2008.09.030
17. Kim S, Lee JH, Choi JY, Kim JM, Kim HS. False-positive rate of a “fourth-generation” HIV antigen/antibody combination assay in an area of low HIV prevalence. Clin Vaccine Immunol. 2010;17(10):1642-1644. doi:10.1128/CVI.00258-10
18. Muthukumar A, Alatoom A, Burns S, et al. Comparison of 4th-generation HIV antigen/antibody combination assay with 3rd-generation HIV antibody assays for the occurrence of false-positive and false-negative results. Lab Med. 2015;46(2):84-e29. doi:10.1309/LMM3X37NSWUCMVRS
19. Mitchell EO, Stewart G, Bajzik O, Ferret M, Bentsen C, Shriver MK. Performance comparison of the 4th generation Bio-Rad Laboratories GS HIV Combo Ag/Ab EIA on the EVOLIS™ automated system versus Abbott ARCHITECT HIV Ag/Ab Combo, Ortho Anti-HIV 1+2 EIA on Vitros ECi and Siemens HIV-1/O/2 enhanced on Advia Centaur. J Clin Virol. 2013;58(suppl 1):e79-e84. doi:10.1016/j.jcv.2013.08.009
20. Dubravac T, Gahan TF, Pentella MA. Use of the Abbott Architect HIV antigen/antibody assay in a low incidence population. J Clin Virol. 2013;58(suppl 1):e76-e78. doi:10.1016/j.jcv.2013.10.020
21. Montesinos I, Eykmans J, Delforge ML. Evaluation of the Bio-Rad Geenius HIV-1/2 test as a confirmatory assay. J Clin Virol. 2014;60(4):399-401. doi:10.1016/j.jcv.2014.04.025
22. van Binsbergen J, Siebelink A, Jacobs A, et al. Improved performance of seroconversion with a 4th generation HIV antigen/antibody assay. J Virol Methods. 1999;82(1):77-84. doi:10.1016/s0166-0934(99)00086-5
23. CLSI. User Protocol for Evaluation of Qualitative Test Performance: Approved Guideline. Second ed. EP12-A2. CLSI; 2008:1-46.
24. Agha Z, Lofgren RP, VanRuiswyk JV, Layde PM. Are patients at Veterans Affairs medical centers sicker? A comparative analysis of health status and medical resource use. Arch Intern Med. 2000;160(21):3252-3257. doi:10.1001/archinte.160.21.3252
25. Eibner C, Krull H, Brown KM, et al. Current and projected characteristics and unique health care needs of the patient population served by the Department of Veterans Affairs. Rand Health Q. 2016;5(4):13. Published 2016 May 9.
26. Morgan RO, Teal CR, Reddy SG, Ford ME, Ashton CM. Measurement in Veterans Affairs Health Services Research: veterans as a special population. Health Serv Res. 2005;40(5, pt 2):1573-1583. doi:10.1111/j.1475-6773.2005.00448.x
27. Nightingale VR, Sher TG, Hansen NB. The impact of receiving an HIV diagnosis and cognitive processing on psychological distress and posttraumatic growth. J Trauma Stress. 2010;23(4):452-460. doi:10.1002/jts.20554
28. Spelman JF, Hunt SC, Seal KH, Burgo-Black AL. Post deployment care for returning combat veterans. J Gen Intern Med. 2012;27(9):1200-1209. doi:10.1007/s11606-012-2061-1
29. Ndase P, Celum C, Kidoguchi L, et al. Frequency of false positive rapid HIV serologic tests in African men and women receiving PrEP for HIV prevention: implications for programmatic roll-out of biomedical interventions. PLoS One. 2015;10(4):e0123005. Published 2015 Apr 17. doi:10.1371/journal.pone.0123005
Ever since the first clinical reports of patients with AIDS in 1981, there have been improvements both in the knowledge base of the pathogenesis of HIV in causing AIDS as well as a progressive refinement in the test methodologies used to diagnose this illness.1-3 Given that there are both public health and clinical benefits in earlier diagnosis and treatment of patients with available antiretroviral therapies, universal screening with opt-out consent has been a standard of practice recommendation by the Centers of Disease Control and Prevention (CDC) since 2006; universal screening with opt-out consent also has been recommended by the US Preventative Task Force and has been widely implemented.4-7
HIV Screening
While HIV screening assays have evolved to be accurate with very high sensitivities and specificities, false-positive results are a significant issue both currently and historically.8-16 The use of an HIV assay on a low prevalence population predictably reduces the positive predictive value (PPV) of even an otherwise accurate assay.8-23 In light of this, laboratory HIV testing algorithms include confirmatory testing to increase the likelihood that the correct diagnosis is being rendered.
The fourth-generation assay has been shown to be more sensitive and specific compared with that of the third-generation assay due to the addition of detection of p24 antigen and the refinement of the antigenic targets for the antibody detection.6,8,11-13,18-20,22 Due to these improvements, in the general population, increased sensitivity/specificity with a reduction in both false positives and false negatives have been reported.
It has been observed in the nonveteran population that switching from the older third-generation to a more sensitive and specific fourth-generation HIV screening assay has reduced the false-positive screening rate.18,19,22 For instance, Muthukumar and colleagues demonstrated a false-positive rate of only 2 out of 99 (2%) tested specimens for the fourth-generation ARCHITECT HIV Ag/Ab Combo assay vs 9 out of 99 tested specimens (9%) for the third-generation ADVIA Centaur HIV 1/O/2 Enhanced assay.18 In addition, it has been noted that fourth-generation HIV screening assays can reduce the window period by detecting HIV infection sooner after initial acute infection.19 Mitchell and colleagues demonstrated even highly specific fourth-generation HIV assays with specificities estimated at 99.7% can have PPVs as low as 25.0% if used in a population of low HIV prevalence (such as a 0.1% prevalence population).19 However, the veteran population has been documented to differ significantly on a number of population variables, including severity of disease and susceptibility to infections, and as a result extrapolation of these data from the general population may be limited.24-26 To our knowledge, this article represents the first study directly examining the reduction in false-positive results with the switch to a fourth-generation HIV generation assay from a third-generation assay for the veteran patient population at a regional US Department of Veterans Affairs (VA) medical center (VAMC).8,11
Methods
Quality assurance documents on test volume were retrospectively reviewed to obtain the number of HIV screening tests that were performed by the laboratory at the Corporal Michael J. Crescenz VAMC (CMJCVAMC) in Philadelphia, Pennsylvania, between March 1, 2016 and February 28, 2017, prior to implementation of the fourth-generation assay. The study also include results from the first year of use of the fourth-generation assay (March 1, 2017 to February 28, 2018). In addition, paper quality assurance records of all positive screening results during those periods were reviewed and manually counted for the abstract presentation of these data.
For assurance of accuracy, a search of all HIV testing assays using Veterans Health Information Systems and Technology Architecture and FileMan also was performed, and the results were compared to records in the Computerized Patient Record System (CPRS). Any discrepancies in the numbers of test results generated by both searches were investigated, and data for the manuscript were derived from records associating tests with particular patients. Only results from patient samples were considered for the electronic search. Quality samples that did not correspond to a true patient as identified in CPRS or same time patient sample duplicates were excluded from the calculations. Basic demographic data (age, ethnicity, and gender) were obtained from this FileMan search. The third-generation assay was the Ortho-Clinical Diagnostics Vitros, and the fourth-generation assay was the Abbott Architect.
To interpret the true HIV result of each sample with a reactive or positive screening result, the CDC laboratory HIV testing algorithm was followed and reviewed with a clinical pathologist or microbiologist director.12,13 All specimens interpreted as HIV positive by the pathologist or microbiologist director were discussed with the clinical health care provider at the time of the test with results added to CPRS after all testing was complete and discussions had taken place. All initially reactive specimens (confirmed with retesting in duplicate on the screening platform with at least 1 repeat reactive result) were further tested with the Bio-Rad Geenius HIV 1/2 Supplemental Assay, which screens for both HIV-1 and HIV-2 antibodies. Specimens with reactive results by this supplemental assay were interpreted as positive for HIV based on the CDC laboratory HIV testing algorithm. Specimens with negative or indeterminant results by the supplemental assay then underwent HIV-1 nucleic acid testing (NAT) using the Roche Diagnostics COBAS AmpliPrep/COBAS TaqMan HIV-1 Test v2.0. Specimens with viral load detected on NAT were positive for HIV infection, while specimens with viral load not detected on NAT testing were interpreted as negative for HIV-1 infection. Although there were no HIV-2 positive or indeterminant specimens during the study period, HIV-2 reactivity also would have been interpreted per the CDC laboratory HIV testing algorithm. Specimens with inadequate volume to complete all testing steps would be interpreted as indeterminant for HIV with request for additional specimen to complete testing. All testing platforms used for HIV testing in the laboratory had been properly validated prior to use.
The number of false positives and indeterminant results was tabulated in Microsoft Excel by month throughout the study period alongside the total number of HIV screening tests performed. Statistical analyses to verify statistical significance was performed by 1-tailed homoscedastic t test calculation using Excel.
Results
From March 1, 2016 to February 28, 2017, 7,516 specimens were screened for HIV, using the third-generation assay, and 52 specimens tested positive for HIV. On further review of these reactive specimens per the CDC laboratory testing algorithm, 24 tests were true positive and 28 were false positives with a PPV of 46% (24/52) (Figure 1).
From March 1, 2017 to February 28, 2018, 7,802 specimens were screened for HIV using a fourth-generation assay and 23 tested positive for HIV. On further review of these reactive specimens per the CDC laboratory testing algorithm, 16 were true positive and 7 were false positives with a PPV of 70% (16/23).
The fourth-generation assay was more specific when compared with the third-generation assay (0.09% vs 0.37%, respectively) with a 75.7% decrease in the false-positivity rate after the implementation of fourth-generation testing. The decreased number of false-positive test results per month with the fourth-generation test implementation was statistically significant (P = .002). The mean (SD) number of false-positive test results for the third-generation assay was 2.3 (1.7) per month, while the fourth-generation assay only had a mean (SD) of 0.58 (0.9) false positives monthly. The decrease in the percentage of false positives per month with the implementation of the fourth-generation assay also was statistically significant (P = .002) (Figure 2).
For population-based reference of the tested population at CMJCVAMC, there was a FileMan search for basic demographic data of patients for the HIV specimens screened by the third- or fourth-generation test (Table). For the population tested by the third-generation assay, 1,114 out of the 7,516 total tested population did not have readily available demographic information by the FileMan search as the specimens originated outside of the facility. For 6,402 of 7,516 patients tested by the third-generation assay with demographic information, the age ranged from 25 to 97 years with a mean of 57 years. This population of 6,402 was 88% male (n = 5,639), 50% African American (n = 3,220) and 43% White (n = 2,756). For the population tested by the fourth-generation assay, 993 of 7,802 total tested population did not have readily available demographic information by the FileMan search as the specimens originated outside of the facility. For the 6,809 of 7,802 patients tested by the fourth-generation assay with demographic information, the age ranged from 24 to 97 years with a mean age of 56 years. This population was 88% male (n = 5,971), 47% African American (n = 3,189), and 46% White (n = 3,149).
Discussion
Current practice guidelines from the CDC and the US Preventive Services Task Force recommend universal screening of the population for HIV infection.5,6 As the general population to be screened would normally have a low prevalence of HIV infection, the risk of a false positive on the initial screen is significant.17 Indeed, the CMJCVAMC experience has been that with the third-generation screening assay, the number of false-positive test results outnumbered the number of true-positive test results. Even with the fourth-generation assay, approximately one-third of the results were false positives. These results are similar to those observed in studies involving nonveteran populations in which the implementation of a fourth-generation screening assay led to significantly fewer false-positive results.18
For laboratories that do not follows CDC testing algorithm guidelines, each false-positive screening result represents a potential opportunity for a HIV misdiagnosis.Even in laboratories with proper procedures in place, false-positive results have consequences for the patients and for the cost-effectiveness of laboratory operations.9-11,18 As per CDC HIV testing guidelines, all positive screening results should be retested, which leads to additional use of technologist time and reagents. After this additional testing is performed and reviewed appropriately, only then can an appropriate final laboratory diagnosis be rendered that meets the standard of laboratory care.
Cost Savings
As observed at CMJCVAMC, the use of a fourth-generation assay with increased sensitivity/specificity led to a reduction in these false-positive results, which improved laboratory efficiency and avoided wasted resources for confirmatory tests.11,18 Cost savings at CMJCVAMC from the implementation of the fourth-generation assay would include technologist time and reagent cost. Generalizable technologist time costs at any institution would include the time needed to perform the confirmatory HIV-1/HIV-2 antibody differentiation assay (slightly less than 1 hour at CMJCVAMC per specimen) and the time needed to perform the viral load assay (about 6 hours to run a batch of 24 tests at CMJCVAMC). We calculated that confirmatory testing cost $184.51 per test at CMJCVAMC. Replacing the third-generation assay with the more sensitive and specific fourth-generation test saved an estimated $3,875 annually. This cost savings does not even consider savings in the pathologist/director’s time for reviewing HIV results after the completion of the algorithm or the clinician/patient costs or anxiety while waiting for results of the confirmatory sequence of tests.
As diagnosis of HIV can have a significant psychological impact on the patient, it is important to ensure the diagnosis conveyed is correct.27 The provision of an HIV diagnosis to a patient has been described as a traumatic stressor capable of causing psychological harm; this harm should ideally be avoided if the HIV diagnosis is not accurate. There can be a temptation, when presented with a positive or reactive screening test that is known to come from an instrument or assay with a very high sensitivity and specificity, to present this result as a diagnosis to the patient. However, a false diagnosis from a false-positive screen would not only be harmful, but given the low prevalence of the disease in the screened population, would happen fairly frequently; in some settings the number of false positives may actually outnumber the number of true positive test results.
Better screening assays with greater specificity (even fractions of a percentage, given that specificities are already > 99%) would help reduce the number of false positives and reduce the number of potential enticements to convey an incorrect diagnosis. Therefore, by adding an additional layer of safety through greater specificity, the fourth-generation assay implementation helped improve the diagnostic safety of the laboratory and reduced the significant error risk to the clinician who would ultimately bear responsibility for conveying the HIV diagnoses to the patient. Given the increased prevalence of psychological and physical ailments in veterans, it may be even more important to ensure the diagnosis is correct to avoid increased psychological harm.27,28
Veteran Population
For the general population, the fourth-generation assay has been shown to be more sensitive and specific when compared with the third-generation assay due to the addition of detection of p24 antigen and the refinement of the antigenic targets for the antibody detection.6,8,11-13,18-20,22 However, the veteran population that receives VA medical care differs significantly from the nonveteran general population. Compared with nonveterans, veterans tend to have generally poorer health status, more comorbid conditions, and greater need to use medical resources.24-26 In addition, veterans also may differ in sociodemographic status, race, ethnicity, and gender.24-26
VA research in the veteran population is unique, and veterans who use VA health care services are an even more highly selected subpopulation.26 Conclusions made from studies of the general population may not always be applicable to the veteran population treated by VA health care services due to these population differences. Therefore, specific studies tailored to this special veteran population in the specific VA health care setting are essential to ensure that the results of the general population truly and definitively apply to the veteran population.
While the false-positive risk is most closely associated with testing in a population of low prevalence, it also should be noted that false-positive screening results also can occur in high-risk individuals, such as an individual on preexposure prophylaxis (PrEP) for continuous behavior that places the individual at high risk of HIV acquisition.8,29 The false-positive result in these cases can lead to a conundrum for the clinician, and the differential diagnosis should consider both detection of very early infection as well as false positive. Interventions could include either stopping PrEP and treating for presumed early primary infection with HIV or continuing the PrEP. These interventions all have the potential to impact the patient whether through the production of resistant HIV virus due to the inadvertent provision of an inadequate treatment regimen, increased risk of infection if taken off PrEP as the patient may likely continue the behavior regardless, or the risks carried by the administration of additional antiretroviral therapies for the complete empiric therapy. Cases of an individual on PrEP who had a false-positive HIV screening test has been reported previously both within and outside the veteran population.8 Better screening tests with greater sensitivity/specificity can only help in guiding better patient care.
Limitations
This quality assurance study was limited to retrospectively identifying the improvement in the false-positive rate on the transition from the third-generation to the more advanced fourth-generation HIV screen. False-positive screen cases could be easily picked up on review of the confirmatory testing per the CDC laboratory HIV testing algorithm.12,13 This study also was a retrospective review of clinically ordered and indicated testing; as a result, without confirmatory testing performed on all negative screen cases, a false-negative rate would not be calculable.
This study also was restricted to only the population being treated in a VA health care setting. This population is known to be different from the general population.24-26
Conclusions
The switch to a fourth-generation assay resulted in a significant reduction in false-positive test results for veteran patients at CMJCVAMC. This reduction in false-positive screening not only reduced laboratory workload due to the necessary confirmatory testing and subsequent review, but also saved costs for technologist’s time and reagents. While this reduction in false-positive results has been documented in nonveteran populations, this is the first study specifically on a veteran population treated at a VAMC.8,11,18 This study confirms previously documented findings of improvement in the false-positive rate of HIV screening tests with the change from third-generation to fourth-generation assay for a veteran population.24
Ever since the first clinical reports of patients with AIDS in 1981, there have been improvements both in the knowledge base of the pathogenesis of HIV in causing AIDS as well as a progressive refinement in the test methodologies used to diagnose this illness.1-3 Given that there are both public health and clinical benefits in earlier diagnosis and treatment of patients with available antiretroviral therapies, universal screening with opt-out consent has been a standard of practice recommendation by the Centers of Disease Control and Prevention (CDC) since 2006; universal screening with opt-out consent also has been recommended by the US Preventative Task Force and has been widely implemented.4-7
HIV Screening
While HIV screening assays have evolved to be accurate with very high sensitivities and specificities, false-positive results are a significant issue both currently and historically.8-16 The use of an HIV assay on a low prevalence population predictably reduces the positive predictive value (PPV) of even an otherwise accurate assay.8-23 In light of this, laboratory HIV testing algorithms include confirmatory testing to increase the likelihood that the correct diagnosis is being rendered.
The fourth-generation assay has been shown to be more sensitive and specific compared with that of the third-generation assay due to the addition of detection of p24 antigen and the refinement of the antigenic targets for the antibody detection.6,8,11-13,18-20,22 Due to these improvements, in the general population, increased sensitivity/specificity with a reduction in both false positives and false negatives have been reported.
It has been observed in the nonveteran population that switching from the older third-generation to a more sensitive and specific fourth-generation HIV screening assay has reduced the false-positive screening rate.18,19,22 For instance, Muthukumar and colleagues demonstrated a false-positive rate of only 2 out of 99 (2%) tested specimens for the fourth-generation ARCHITECT HIV Ag/Ab Combo assay vs 9 out of 99 tested specimens (9%) for the third-generation ADVIA Centaur HIV 1/O/2 Enhanced assay.18 In addition, it has been noted that fourth-generation HIV screening assays can reduce the window period by detecting HIV infection sooner after initial acute infection.19 Mitchell and colleagues demonstrated even highly specific fourth-generation HIV assays with specificities estimated at 99.7% can have PPVs as low as 25.0% if used in a population of low HIV prevalence (such as a 0.1% prevalence population).19 However, the veteran population has been documented to differ significantly on a number of population variables, including severity of disease and susceptibility to infections, and as a result extrapolation of these data from the general population may be limited.24-26 To our knowledge, this article represents the first study directly examining the reduction in false-positive results with the switch to a fourth-generation HIV generation assay from a third-generation assay for the veteran patient population at a regional US Department of Veterans Affairs (VA) medical center (VAMC).8,11
Methods
Quality assurance documents on test volume were retrospectively reviewed to obtain the number of HIV screening tests that were performed by the laboratory at the Corporal Michael J. Crescenz VAMC (CMJCVAMC) in Philadelphia, Pennsylvania, between March 1, 2016 and February 28, 2017, prior to implementation of the fourth-generation assay. The study also include results from the first year of use of the fourth-generation assay (March 1, 2017 to February 28, 2018). In addition, paper quality assurance records of all positive screening results during those periods were reviewed and manually counted for the abstract presentation of these data.
For assurance of accuracy, a search of all HIV testing assays using Veterans Health Information Systems and Technology Architecture and FileMan also was performed, and the results were compared to records in the Computerized Patient Record System (CPRS). Any discrepancies in the numbers of test results generated by both searches were investigated, and data for the manuscript were derived from records associating tests with particular patients. Only results from patient samples were considered for the electronic search. Quality samples that did not correspond to a true patient as identified in CPRS or same time patient sample duplicates were excluded from the calculations. Basic demographic data (age, ethnicity, and gender) were obtained from this FileMan search. The third-generation assay was the Ortho-Clinical Diagnostics Vitros, and the fourth-generation assay was the Abbott Architect.
To interpret the true HIV result of each sample with a reactive or positive screening result, the CDC laboratory HIV testing algorithm was followed and reviewed with a clinical pathologist or microbiologist director.12,13 All specimens interpreted as HIV positive by the pathologist or microbiologist director were discussed with the clinical health care provider at the time of the test with results added to CPRS after all testing was complete and discussions had taken place. All initially reactive specimens (confirmed with retesting in duplicate on the screening platform with at least 1 repeat reactive result) were further tested with the Bio-Rad Geenius HIV 1/2 Supplemental Assay, which screens for both HIV-1 and HIV-2 antibodies. Specimens with reactive results by this supplemental assay were interpreted as positive for HIV based on the CDC laboratory HIV testing algorithm. Specimens with negative or indeterminant results by the supplemental assay then underwent HIV-1 nucleic acid testing (NAT) using the Roche Diagnostics COBAS AmpliPrep/COBAS TaqMan HIV-1 Test v2.0. Specimens with viral load detected on NAT were positive for HIV infection, while specimens with viral load not detected on NAT testing were interpreted as negative for HIV-1 infection. Although there were no HIV-2 positive or indeterminant specimens during the study period, HIV-2 reactivity also would have been interpreted per the CDC laboratory HIV testing algorithm. Specimens with inadequate volume to complete all testing steps would be interpreted as indeterminant for HIV with request for additional specimen to complete testing. All testing platforms used for HIV testing in the laboratory had been properly validated prior to use.
The number of false positives and indeterminant results was tabulated in Microsoft Excel by month throughout the study period alongside the total number of HIV screening tests performed. Statistical analyses to verify statistical significance was performed by 1-tailed homoscedastic t test calculation using Excel.
Results
From March 1, 2016 to February 28, 2017, 7,516 specimens were screened for HIV, using the third-generation assay, and 52 specimens tested positive for HIV. On further review of these reactive specimens per the CDC laboratory testing algorithm, 24 tests were true positive and 28 were false positives with a PPV of 46% (24/52) (Figure 1).
From March 1, 2017 to February 28, 2018, 7,802 specimens were screened for HIV using a fourth-generation assay and 23 tested positive for HIV. On further review of these reactive specimens per the CDC laboratory testing algorithm, 16 were true positive and 7 were false positives with a PPV of 70% (16/23).
The fourth-generation assay was more specific when compared with the third-generation assay (0.09% vs 0.37%, respectively) with a 75.7% decrease in the false-positivity rate after the implementation of fourth-generation testing. The decreased number of false-positive test results per month with the fourth-generation test implementation was statistically significant (P = .002). The mean (SD) number of false-positive test results for the third-generation assay was 2.3 (1.7) per month, while the fourth-generation assay only had a mean (SD) of 0.58 (0.9) false positives monthly. The decrease in the percentage of false positives per month with the implementation of the fourth-generation assay also was statistically significant (P = .002) (Figure 2).
For population-based reference of the tested population at CMJCVAMC, there was a FileMan search for basic demographic data of patients for the HIV specimens screened by the third- or fourth-generation test (Table). For the population tested by the third-generation assay, 1,114 out of the 7,516 total tested population did not have readily available demographic information by the FileMan search as the specimens originated outside of the facility. For 6,402 of 7,516 patients tested by the third-generation assay with demographic information, the age ranged from 25 to 97 years with a mean of 57 years. This population of 6,402 was 88% male (n = 5,639), 50% African American (n = 3,220) and 43% White (n = 2,756). For the population tested by the fourth-generation assay, 993 of 7,802 total tested population did not have readily available demographic information by the FileMan search as the specimens originated outside of the facility. For the 6,809 of 7,802 patients tested by the fourth-generation assay with demographic information, the age ranged from 24 to 97 years with a mean age of 56 years. This population was 88% male (n = 5,971), 47% African American (n = 3,189), and 46% White (n = 3,149).
Discussion
Current practice guidelines from the CDC and the US Preventive Services Task Force recommend universal screening of the population for HIV infection.5,6 As the general population to be screened would normally have a low prevalence of HIV infection, the risk of a false positive on the initial screen is significant.17 Indeed, the CMJCVAMC experience has been that with the third-generation screening assay, the number of false-positive test results outnumbered the number of true-positive test results. Even with the fourth-generation assay, approximately one-third of the results were false positives. These results are similar to those observed in studies involving nonveteran populations in which the implementation of a fourth-generation screening assay led to significantly fewer false-positive results.18
For laboratories that do not follows CDC testing algorithm guidelines, each false-positive screening result represents a potential opportunity for a HIV misdiagnosis.Even in laboratories with proper procedures in place, false-positive results have consequences for the patients and for the cost-effectiveness of laboratory operations.9-11,18 As per CDC HIV testing guidelines, all positive screening results should be retested, which leads to additional use of technologist time and reagents. After this additional testing is performed and reviewed appropriately, only then can an appropriate final laboratory diagnosis be rendered that meets the standard of laboratory care.
Cost Savings
As observed at CMJCVAMC, the use of a fourth-generation assay with increased sensitivity/specificity led to a reduction in these false-positive results, which improved laboratory efficiency and avoided wasted resources for confirmatory tests.11,18 Cost savings at CMJCVAMC from the implementation of the fourth-generation assay would include technologist time and reagent cost. Generalizable technologist time costs at any institution would include the time needed to perform the confirmatory HIV-1/HIV-2 antibody differentiation assay (slightly less than 1 hour at CMJCVAMC per specimen) and the time needed to perform the viral load assay (about 6 hours to run a batch of 24 tests at CMJCVAMC). We calculated that confirmatory testing cost $184.51 per test at CMJCVAMC. Replacing the third-generation assay with the more sensitive and specific fourth-generation test saved an estimated $3,875 annually. This cost savings does not even consider savings in the pathologist/director’s time for reviewing HIV results after the completion of the algorithm or the clinician/patient costs or anxiety while waiting for results of the confirmatory sequence of tests.
As diagnosis of HIV can have a significant psychological impact on the patient, it is important to ensure the diagnosis conveyed is correct.27 The provision of an HIV diagnosis to a patient has been described as a traumatic stressor capable of causing psychological harm; this harm should ideally be avoided if the HIV diagnosis is not accurate. There can be a temptation, when presented with a positive or reactive screening test that is known to come from an instrument or assay with a very high sensitivity and specificity, to present this result as a diagnosis to the patient. However, a false diagnosis from a false-positive screen would not only be harmful, but given the low prevalence of the disease in the screened population, would happen fairly frequently; in some settings the number of false positives may actually outnumber the number of true positive test results.
Better screening assays with greater specificity (even fractions of a percentage, given that specificities are already > 99%) would help reduce the number of false positives and reduce the number of potential enticements to convey an incorrect diagnosis. Therefore, by adding an additional layer of safety through greater specificity, the fourth-generation assay implementation helped improve the diagnostic safety of the laboratory and reduced the significant error risk to the clinician who would ultimately bear responsibility for conveying the HIV diagnoses to the patient. Given the increased prevalence of psychological and physical ailments in veterans, it may be even more important to ensure the diagnosis is correct to avoid increased psychological harm.27,28
Veteran Population
For the general population, the fourth-generation assay has been shown to be more sensitive and specific when compared with the third-generation assay due to the addition of detection of p24 antigen and the refinement of the antigenic targets for the antibody detection.6,8,11-13,18-20,22 However, the veteran population that receives VA medical care differs significantly from the nonveteran general population. Compared with nonveterans, veterans tend to have generally poorer health status, more comorbid conditions, and greater need to use medical resources.24-26 In addition, veterans also may differ in sociodemographic status, race, ethnicity, and gender.24-26
VA research in the veteran population is unique, and veterans who use VA health care services are an even more highly selected subpopulation.26 Conclusions made from studies of the general population may not always be applicable to the veteran population treated by VA health care services due to these population differences. Therefore, specific studies tailored to this special veteran population in the specific VA health care setting are essential to ensure that the results of the general population truly and definitively apply to the veteran population.
While the false-positive risk is most closely associated with testing in a population of low prevalence, it also should be noted that false-positive screening results also can occur in high-risk individuals, such as an individual on preexposure prophylaxis (PrEP) for continuous behavior that places the individual at high risk of HIV acquisition.8,29 The false-positive result in these cases can lead to a conundrum for the clinician, and the differential diagnosis should consider both detection of very early infection as well as false positive. Interventions could include either stopping PrEP and treating for presumed early primary infection with HIV or continuing the PrEP. These interventions all have the potential to impact the patient whether through the production of resistant HIV virus due to the inadvertent provision of an inadequate treatment regimen, increased risk of infection if taken off PrEP as the patient may likely continue the behavior regardless, or the risks carried by the administration of additional antiretroviral therapies for the complete empiric therapy. Cases of an individual on PrEP who had a false-positive HIV screening test has been reported previously both within and outside the veteran population.8 Better screening tests with greater sensitivity/specificity can only help in guiding better patient care.
Limitations
This quality assurance study was limited to retrospectively identifying the improvement in the false-positive rate on the transition from the third-generation to the more advanced fourth-generation HIV screen. False-positive screen cases could be easily picked up on review of the confirmatory testing per the CDC laboratory HIV testing algorithm.12,13 This study also was a retrospective review of clinically ordered and indicated testing; as a result, without confirmatory testing performed on all negative screen cases, a false-negative rate would not be calculable.
This study also was restricted to only the population being treated in a VA health care setting. This population is known to be different from the general population.24-26
Conclusions
The switch to a fourth-generation assay resulted in a significant reduction in false-positive test results for veteran patients at CMJCVAMC. This reduction in false-positive screening not only reduced laboratory workload due to the necessary confirmatory testing and subsequent review, but also saved costs for technologist’s time and reagents. While this reduction in false-positive results has been documented in nonveteran populations, this is the first study specifically on a veteran population treated at a VAMC.8,11,18 This study confirms previously documented findings of improvement in the false-positive rate of HIV screening tests with the change from third-generation to fourth-generation assay for a veteran population.24
1. Feinberg MB. Changing the natural history of HIV disease. Lancet. 1996;348(9022):239-246. doi:10.1016/s0140-6736(96)06231-9.
2. Alexander TS. Human immunodeficiency virus diagnostic testing: 30 years of evolution. Clin Vaccine Immunol. 2016;23(4):249-253. Published 2016 Apr 4. doi:10.1128/CVI.00053-16
3. Mortimer PP, Parry JV, Mortimer JY. Which anti-HTLV III/LAV assays for screening and confirmatory testing?. Lancet. 1985;2(8460):873-877. doi:10.1016/s0140-6736(85)90136-9
4. Holmberg SD, Palella FJ Jr, Lichtenstein KA, Havlir DV. The case for earlier treatment of HIV infection [published correction appears in Clin Infect Dis. 2004 Dec 15;39(12):1869]. Clin Infect Dis. 2004;39(11):1699-1704. doi:10.1086/425743
5. US Preventive Services Task Force, Owens DK, Davidson KW, et al. Screening for HIV Infection: US Preventive Services Task Force Recommendation Statement. JAMA. 2019;321(23):2326-2336. doi:10.1001/jama.2019.6587
6. Branson BM, Handsfield HH, Lampe MA, et al. Revised recommendations for HIV testing of adults, adolescents, and pregnant women in health-care settings. MMWR Recomm Rep. 2006;55(RR-14):1-CE4.
7. Bayer R, Philbin M, Remien RH. The end of written informed consent for HIV testing: not with a bang but a whimper. Am J Public Health. 2017;107(8):1259-1265. doi:10.2105/AJPH.2017.303819
8. Petersen J, Jhala D. Its not HIV! The pitfall of unconfirmed positive HIV screening assays. Abstract presented at: Annual Meeting Pennsylvania Association of Pathologists; April 14, 2018.
9. Wood RW, Dunphy C, Okita K, Swenson P. Two “HIV-infected” persons not really infected. Arch Intern Med. 2003;163(15):1857-1859. doi:10.1001/archinte.163.15.1857
10. Permpalung N, Ungprasert P, Chongnarungsin D, Okoli A, Hyman CL. A diagnostic blind spot: acute infectious mononucleosis or acute retroviral syndrome. Am J Med. 2013;126(9):e5-e6. doi:10.1016/j.amjmed.2013.03.017
11. Dalal S, Petersen J, Luta D, Jhala D. Third- to fourth-generation HIV testing: reduction in false-positive results with the new way of testing, the Corporal Michael J. Crescenz Veteran Affairs Medical Center (CMCVAMC) Experience. Am J Clin Pathol.2018;150(suppl 1):S70-S71. doi:10.1093/ajcp/aqy093.172
12. Centers for Disease Control and Prevention. Laboratory testing for the diagnosis of HIV infection: updated recommendations. Published June 27, 2014. Accessed April 14, 2021. doi:10.15620/cdc.23447
13. Centers for Disease Control and Prevention. 2018 quick reference guide: recommended laboratory HIV testing algorithm for serum or plasma specimens. Updated January 2018. Accessed April 14, 202. https://stacks.cdc.gov/view/cdc/50872
14. Masciotra S, McDougal JS, Feldman J, Sprinkle P, Wesolowski L, Owen SM. Evaluation of an alternative HIV diagnostic algorithm using specimens from seroconversion panels and persons with established HIV infections. J Clin Virol. 2011;52(suppl 1):S17-S22. doi:10.1016/j.jcv.2011.09.011
15. Morton A. When lab tests lie … heterophile antibodies. Aust Fam Physician. 2014;43(6):391-393.
16. Spencer DV, Nolte FS, Zhu Y. Heterophilic antibody interference causing false-positive rapid human immunodeficiency virus antibody testing. Clin Chim Acta. 2009;399(1-2):121-122. doi:10.1016/j.cca.2008.09.030
17. Kim S, Lee JH, Choi JY, Kim JM, Kim HS. False-positive rate of a “fourth-generation” HIV antigen/antibody combination assay in an area of low HIV prevalence. Clin Vaccine Immunol. 2010;17(10):1642-1644. doi:10.1128/CVI.00258-10
18. Muthukumar A, Alatoom A, Burns S, et al. Comparison of 4th-generation HIV antigen/antibody combination assay with 3rd-generation HIV antibody assays for the occurrence of false-positive and false-negative results. Lab Med. 2015;46(2):84-e29. doi:10.1309/LMM3X37NSWUCMVRS
19. Mitchell EO, Stewart G, Bajzik O, Ferret M, Bentsen C, Shriver MK. Performance comparison of the 4th generation Bio-Rad Laboratories GS HIV Combo Ag/Ab EIA on the EVOLIS™ automated system versus Abbott ARCHITECT HIV Ag/Ab Combo, Ortho Anti-HIV 1+2 EIA on Vitros ECi and Siemens HIV-1/O/2 enhanced on Advia Centaur. J Clin Virol. 2013;58(suppl 1):e79-e84. doi:10.1016/j.jcv.2013.08.009
20. Dubravac T, Gahan TF, Pentella MA. Use of the Abbott Architect HIV antigen/antibody assay in a low incidence population. J Clin Virol. 2013;58(suppl 1):e76-e78. doi:10.1016/j.jcv.2013.10.020
21. Montesinos I, Eykmans J, Delforge ML. Evaluation of the Bio-Rad Geenius HIV-1/2 test as a confirmatory assay. J Clin Virol. 2014;60(4):399-401. doi:10.1016/j.jcv.2014.04.025
22. van Binsbergen J, Siebelink A, Jacobs A, et al. Improved performance of seroconversion with a 4th generation HIV antigen/antibody assay. J Virol Methods. 1999;82(1):77-84. doi:10.1016/s0166-0934(99)00086-5
23. CLSI. User Protocol for Evaluation of Qualitative Test Performance: Approved Guideline. Second ed. EP12-A2. CLSI; 2008:1-46.
24. Agha Z, Lofgren RP, VanRuiswyk JV, Layde PM. Are patients at Veterans Affairs medical centers sicker? A comparative analysis of health status and medical resource use. Arch Intern Med. 2000;160(21):3252-3257. doi:10.1001/archinte.160.21.3252
25. Eibner C, Krull H, Brown KM, et al. Current and projected characteristics and unique health care needs of the patient population served by the Department of Veterans Affairs. Rand Health Q. 2016;5(4):13. Published 2016 May 9.
26. Morgan RO, Teal CR, Reddy SG, Ford ME, Ashton CM. Measurement in Veterans Affairs Health Services Research: veterans as a special population. Health Serv Res. 2005;40(5, pt 2):1573-1583. doi:10.1111/j.1475-6773.2005.00448.x
27. Nightingale VR, Sher TG, Hansen NB. The impact of receiving an HIV diagnosis and cognitive processing on psychological distress and posttraumatic growth. J Trauma Stress. 2010;23(4):452-460. doi:10.1002/jts.20554
28. Spelman JF, Hunt SC, Seal KH, Burgo-Black AL. Post deployment care for returning combat veterans. J Gen Intern Med. 2012;27(9):1200-1209. doi:10.1007/s11606-012-2061-1
29. Ndase P, Celum C, Kidoguchi L, et al. Frequency of false positive rapid HIV serologic tests in African men and women receiving PrEP for HIV prevention: implications for programmatic roll-out of biomedical interventions. PLoS One. 2015;10(4):e0123005. Published 2015 Apr 17. doi:10.1371/journal.pone.0123005
1. Feinberg MB. Changing the natural history of HIV disease. Lancet. 1996;348(9022):239-246. doi:10.1016/s0140-6736(96)06231-9.
2. Alexander TS. Human immunodeficiency virus diagnostic testing: 30 years of evolution. Clin Vaccine Immunol. 2016;23(4):249-253. Published 2016 Apr 4. doi:10.1128/CVI.00053-16
3. Mortimer PP, Parry JV, Mortimer JY. Which anti-HTLV III/LAV assays for screening and confirmatory testing?. Lancet. 1985;2(8460):873-877. doi:10.1016/s0140-6736(85)90136-9
4. Holmberg SD, Palella FJ Jr, Lichtenstein KA, Havlir DV. The case for earlier treatment of HIV infection [published correction appears in Clin Infect Dis. 2004 Dec 15;39(12):1869]. Clin Infect Dis. 2004;39(11):1699-1704. doi:10.1086/425743
5. US Preventive Services Task Force, Owens DK, Davidson KW, et al. Screening for HIV Infection: US Preventive Services Task Force Recommendation Statement. JAMA. 2019;321(23):2326-2336. doi:10.1001/jama.2019.6587
6. Branson BM, Handsfield HH, Lampe MA, et al. Revised recommendations for HIV testing of adults, adolescents, and pregnant women in health-care settings. MMWR Recomm Rep. 2006;55(RR-14):1-CE4.
7. Bayer R, Philbin M, Remien RH. The end of written informed consent for HIV testing: not with a bang but a whimper. Am J Public Health. 2017;107(8):1259-1265. doi:10.2105/AJPH.2017.303819
8. Petersen J, Jhala D. Its not HIV! The pitfall of unconfirmed positive HIV screening assays. Abstract presented at: Annual Meeting Pennsylvania Association of Pathologists; April 14, 2018.
9. Wood RW, Dunphy C, Okita K, Swenson P. Two “HIV-infected” persons not really infected. Arch Intern Med. 2003;163(15):1857-1859. doi:10.1001/archinte.163.15.1857
10. Permpalung N, Ungprasert P, Chongnarungsin D, Okoli A, Hyman CL. A diagnostic blind spot: acute infectious mononucleosis or acute retroviral syndrome. Am J Med. 2013;126(9):e5-e6. doi:10.1016/j.amjmed.2013.03.017
11. Dalal S, Petersen J, Luta D, Jhala D. Third- to fourth-generation HIV testing: reduction in false-positive results with the new way of testing, the Corporal Michael J. Crescenz Veteran Affairs Medical Center (CMCVAMC) Experience. Am J Clin Pathol.2018;150(suppl 1):S70-S71. doi:10.1093/ajcp/aqy093.172
12. Centers for Disease Control and Prevention. Laboratory testing for the diagnosis of HIV infection: updated recommendations. Published June 27, 2014. Accessed April 14, 2021. doi:10.15620/cdc.23447
13. Centers for Disease Control and Prevention. 2018 quick reference guide: recommended laboratory HIV testing algorithm for serum or plasma specimens. Updated January 2018. Accessed April 14, 202. https://stacks.cdc.gov/view/cdc/50872
14. Masciotra S, McDougal JS, Feldman J, Sprinkle P, Wesolowski L, Owen SM. Evaluation of an alternative HIV diagnostic algorithm using specimens from seroconversion panels and persons with established HIV infections. J Clin Virol. 2011;52(suppl 1):S17-S22. doi:10.1016/j.jcv.2011.09.011
15. Morton A. When lab tests lie … heterophile antibodies. Aust Fam Physician. 2014;43(6):391-393.
16. Spencer DV, Nolte FS, Zhu Y. Heterophilic antibody interference causing false-positive rapid human immunodeficiency virus antibody testing. Clin Chim Acta. 2009;399(1-2):121-122. doi:10.1016/j.cca.2008.09.030
17. Kim S, Lee JH, Choi JY, Kim JM, Kim HS. False-positive rate of a “fourth-generation” HIV antigen/antibody combination assay in an area of low HIV prevalence. Clin Vaccine Immunol. 2010;17(10):1642-1644. doi:10.1128/CVI.00258-10
18. Muthukumar A, Alatoom A, Burns S, et al. Comparison of 4th-generation HIV antigen/antibody combination assay with 3rd-generation HIV antibody assays for the occurrence of false-positive and false-negative results. Lab Med. 2015;46(2):84-e29. doi:10.1309/LMM3X37NSWUCMVRS
19. Mitchell EO, Stewart G, Bajzik O, Ferret M, Bentsen C, Shriver MK. Performance comparison of the 4th generation Bio-Rad Laboratories GS HIV Combo Ag/Ab EIA on the EVOLIS™ automated system versus Abbott ARCHITECT HIV Ag/Ab Combo, Ortho Anti-HIV 1+2 EIA on Vitros ECi and Siemens HIV-1/O/2 enhanced on Advia Centaur. J Clin Virol. 2013;58(suppl 1):e79-e84. doi:10.1016/j.jcv.2013.08.009
20. Dubravac T, Gahan TF, Pentella MA. Use of the Abbott Architect HIV antigen/antibody assay in a low incidence population. J Clin Virol. 2013;58(suppl 1):e76-e78. doi:10.1016/j.jcv.2013.10.020
21. Montesinos I, Eykmans J, Delforge ML. Evaluation of the Bio-Rad Geenius HIV-1/2 test as a confirmatory assay. J Clin Virol. 2014;60(4):399-401. doi:10.1016/j.jcv.2014.04.025
22. van Binsbergen J, Siebelink A, Jacobs A, et al. Improved performance of seroconversion with a 4th generation HIV antigen/antibody assay. J Virol Methods. 1999;82(1):77-84. doi:10.1016/s0166-0934(99)00086-5
23. CLSI. User Protocol for Evaluation of Qualitative Test Performance: Approved Guideline. Second ed. EP12-A2. CLSI; 2008:1-46.
24. Agha Z, Lofgren RP, VanRuiswyk JV, Layde PM. Are patients at Veterans Affairs medical centers sicker? A comparative analysis of health status and medical resource use. Arch Intern Med. 2000;160(21):3252-3257. doi:10.1001/archinte.160.21.3252
25. Eibner C, Krull H, Brown KM, et al. Current and projected characteristics and unique health care needs of the patient population served by the Department of Veterans Affairs. Rand Health Q. 2016;5(4):13. Published 2016 May 9.
26. Morgan RO, Teal CR, Reddy SG, Ford ME, Ashton CM. Measurement in Veterans Affairs Health Services Research: veterans as a special population. Health Serv Res. 2005;40(5, pt 2):1573-1583. doi:10.1111/j.1475-6773.2005.00448.x
27. Nightingale VR, Sher TG, Hansen NB. The impact of receiving an HIV diagnosis and cognitive processing on psychological distress and posttraumatic growth. J Trauma Stress. 2010;23(4):452-460. doi:10.1002/jts.20554
28. Spelman JF, Hunt SC, Seal KH, Burgo-Black AL. Post deployment care for returning combat veterans. J Gen Intern Med. 2012;27(9):1200-1209. doi:10.1007/s11606-012-2061-1
29. Ndase P, Celum C, Kidoguchi L, et al. Frequency of false positive rapid HIV serologic tests in African men and women receiving PrEP for HIV prevention: implications for programmatic roll-out of biomedical interventions. PLoS One. 2015;10(4):e0123005. Published 2015 Apr 17. doi:10.1371/journal.pone.0123005
Liquid Biopsies in a Veteran Patient Population With Advanced Prostate and Lung Non-Small Cell Carcinomas: A New Paradigm and Unique Challenge in Personalized Medicine
The advent of liquid biopsies targeting genetic mutations in solid tumors is a major milestone in the field of precision oncology.1 Conventional methods of obtaining tissue for molecular studies are limited by sample size and often do not represent the entire bulk of the tumor.2 This newer minimally invasive, revolutionary technique analyzes circulating cell-free DNA carrying tumor-specific alterations (circulating tumor DNA [ctDNA]) in peripheral blood and detects signature genomic alterations.1 Tp53 mutations have been reported in 25 to 40% of prostatic cancers and > 50% of non-small cell lung cancers (NSCLC), being more common in late-stage and hormone refractory prostate cancers.3,4 Tp53 mutation has been found to be associated with poor prognosis and increased germline mutations.5
The veteran patient population has distinct demographic characteristics that make veterans more vulnerable to genetic mutations and malignancies, including risk of exposure to Agent Orange, smoking, substance abuse, and asbestos. This area is understudied and extremely sparse in the literature for frequency of genetic mutations, risk factors in solid malignancies occurring in the veteran patient population, and the clinical impact of these risk factors. We herein present a quality assurance study for the utility of liquid biopsies regarding the frequency of DNA damage repair (DDR) gene, Tp53, and androgen receptor (AR) mutations. The clinical impact in advanced lung and prostate cancers in the veteran patient population and frequency are the quality assurance observations that are the study endpoints.
Methods
We reviewed for quality assurance documentation from the Foundation Medicine (www.foundationmedicine.com) cancer biomarker tests on liquid biopsies performed at the Corporal Michael J. Crescenz Veteran Affairs Medical Center in Philadelphia, Pennsylvania from May 2019 to April 15, 2020. All biopsies were performed on cancers with biochemical, imaging or tissue evidence of advanced tumor progression. The testing was performed on advanced solid malignancies, including NSCLC, prostate adenocarcinoma, and metastatic colon cancer. Statistical data for adequacy; cases with notable mutations; frequency; and type of mutations of AR, DDR, and Tp53 were noted. General and specific risk factors associated with the veteran patient population were studied and matched with the type of mutations (Table 1).
Results
Thirty-one liquid biopsies were performed over this period—23 for prostate cancer, 7 for patients with lung cancer patients, and 1 for a patient with colon cancer. Of 31 cases, sensitivity/adequacy of liquid biopsy for genetic mutation was detected in 29 (93.5%) cases (Figure 1). Two inadequate biopsies (both from patients with prostate cancer) were excluded from the study, leaving 29 liquid biopsies with adequate ctDNA for analysis that were considered for further statistical purpose—21 prostate, 7 lung, and 1 colon cancer.
Multiple (common and different) genetic mutations were identified; however, our study subcategorized the mutations into the those that were related to prostate cancer, lung cancer, and some common mutations that occur in both cancers. Only the significant ones will be discussed in this review and equivocal result for AR is excluded from this study. Of the 21 prostate cancers, 4 (19.0%) had directed the targeted therapy to driver mutation (AR being most common in prostate cancer), while KRAS mutation, which was more common in lung cancer, was detected in 2/7 (28.6%) lung cancers. Mutations common to both cancer types were DDR gene mutations, which is a broad name for numerous genes including CDK12, ATM, and CHEK2.
Of all cases irrespective of the cancer type, 23/29 (79.3%) showed notable mutations. DDR gene mutations were found in 6 of 21 (28.5%) patients with prostate cancer and 8 of 23 (34.7%) patients with advanced prostate and lung cancers, indicating poor outcome and possible resistance to the current therapy. Of 23 patients showing mutations irrespective of the cancer type, 15 (65.2%) harbored Tp53 mutations, which is much more frequent in veteran patient population when compared with the literature. Fifteen of the 31 (48.4%) total patients were Vietnam War-era veterans who were potentially exposed to Agent Orange and 20 (64.5%) patients who were not Vietnam War-era veterans had a history that included smoking (Figure 2).
Discussion
The veteran patient population is a unique cohort due to its distinct demographic characteristics with a high volume of cancer cases diagnosed each year. According to data from VA Central Cancer Registry (VACCR), the most frequently diagnosed cancers are prostate (29%) and lung (18%).6
Liquid biopsy is a novel, promising technology that uses ctDNA and circulating tumor cells in peripheral blood for detecting genetic alterations through next generation sequencing.7-9 The advent of this minimally invasive, revolutionary technology has been a breakthrough in the field of precision oncology for prognosis, to monitor treatment response or resistance to therapy and further personalize cancer therapy.9,10
Comprehensive genomic profiling by liquid biopsy has many advantages over the molecular studies performed on tissue biopsy. Due to the tumor heterogeneity, tissue samples may not represent the full profile of the tumor genomics of cancer, while liquid biopsy has full presentation of the disease.11,12 Many times, tissue biopsy may be limited by a sample size that precludes full genetic profiling in addition to higher total cost, potential technical issues during processing, and possible side effects of the biopsy procedure.7,13 Additionally, as the tumor progresses, new driver mutations other than the ones previously detected on the primary tissue may emerge, which can confer resistance to the existing therapy.7,13
Advanced prostatic and lung carcinomas with biochemical, distant organ, or bony progression harbor unique signature genetic mutations indicating poor prognosis, lack of response or resistance to the existing therapy, and high risk of relapse.14,15 Some of the unique characteristics of the veteran patient population include a more aged patient population multiple comorbidities, higher frequency of > 1 type of cancer, advanced cancer stage at presentation, and specific risks factors such as exposure to Agent Orange in veterans who served during the Vietnam War era.16,17 We studied the utility of liquid biopsy in cancer care, including type and incidence of genomic alterations associated with advanced prostate and lung cancers, in this unique patient population.
The amount of cell-free DNA (cfDNA), also known as ctDNA varies widely in cancer patients. Some of the factors associated with low concentration of cfDNA are disease stage, intervening therapy, proliferation rates, and tumor vascularization.18,19 In the peripheral blood, of the total cfDNA, fractions of cfDNA varies from 0.01 to 90%.18,19 All samples containing ≥ 20 ng cfDNA (20 - 100 ng) were subjected to the hybrid capture-based NGS FoundationACT assay.20 In our study, 2 specimens did not meet the minimum criteria of adequacy (20 ng cfDNA); however, the overall adequacy rate for the detection of mutation, irrespective of the cancer type was 29 of 31 (93.5%) with only 2 inadequate samples. This rate is higher than the rate reported in the literature, which is about 70%.20
Significant differences were encountered in the incidence of DNA damage repair genes including Tp53 mutations when compared with those in the general patient population (Table 2). According to recent National Comprehensive Cancer Network (NCCN) guidelines, all prostate cancers should be screened for DDR gene mutations as these genes are common in aggressive prostate cancers and strongly associated with poor outcomes and shortened survival. Due to relatively high frequency of DDR gene mutations in advanced prostatic cancers, liquid biopsy in patients with these advanced stage prostate cancers may be a useful tool in clinical decision making and exploring targeted therapy.20
Mutations in BRCA2, ATM, CDK12, and CHEK2 (DDR gene family) are common. Incidence of ATM and CDK12 mutations in the literature is 3 to 6% of cases.21 Of 21 liquid biopsies of advanced prostate cancer patients, we found combined DDR gene mutation of ATM, CHEK2, and CDK12 genes in 6 (28.5%) cases, which is substantially higher than the 3 to 6% rate reported in the literature.21-24 Of the 23 patients who had notable mutations in our liquid biopsies, including both advanced prostate and lung cancer cases, 8 (34.7%) also showed mutation of the genes of DDR family. Our study did not show BRCA2 mutation, which is otherwise common in the literature.
We also evaluated the frequency of the most commonly occurring genetic mutations, Tp53 in advanced solid malignancies, especially advanced prostate and NSCLC. Previous studies have reported Tp53 mutation in association with risk factors (carcinogens) of cancer and have been a surrogate marker of poor survival or lack of response of therapy.25 Knowledge of Tp53 mutation is crucial for closer disease monitoring, preparing the patient for rapid progression, and encouraging the physician to prepare future lines of therapy.25-27 Although Tp53 mutation varies with histologic type and tissue of origin, Beltran and colleagues reported it in 30 to 40% of tumors, while Robles and colleagues reported about 40 to 42% incidence.25,27
Our study showed notable mutations in 23 of 29 adequate cases. Further, our study showed a high frequency of mutated Tp53 in 65.2% of combined advanced prostate and NSCLC cases. We then correlated cases of Vietnam War-era veterans with risk potential of Agent Orange exposure and Tp53 mutation. We found 7 of 15 Vietnam War-era veterans were positive for Tp53 mutations irrespective of the cancer type. The high incidence of Tp53 mutations in advanced prostate and lung carcinomas in the veteran patient population makes this tumor marker an aspiration not only as a surrogate of aggressive disease and tumor progression, but also as a key marker for targeted therapy in advanced prostate and lung cancers with loss of Tp53 function (Figure 3).
Mutations and amplifications in the AR gene are fundamental to progression of prostate cancer associated with advanced, hormone-refractory prostate cancer with the potential for targeted therapy with AR inhibitors. In our study, AR amplification was detected in 4 of 21 (19%) advanced prostate cancer cases, which is significantly lower than the 30 to 50% previously reported in the literature.28-32 Neither AR amplification or mutation was noted in advanced NSCLC in our study as previously reported in literature by Brennan and colleagues and Wang and colleagues.33-35 This is significant as it provides a pathway for future studies to focus on additional driver mutations for targeted therapies in advanced prostate carcinoma. To date, AR gene mutation does not play a role for personalized therapy in advanced NSCLC. Perhaps, a large cohort study with longitudinal analysis is needed for absolutely ruling out the possibility of personalized medicine in advanced lung cancer using this biomarker.
Conclusions
Liquid biopsy successfully provides precision-based oncology and information for decision making in this unique population of veterans. Difference in frequency of the genetic mutations in this cohort can provide future insight into disease progression, lack of response, and mechanism of resistance to the implemented therapy. Future studies focused on this veteran patient population are needed for developing targeted therapies and patient tailored oncologic therapy. ctDNA has a high potential for monitoring clinically relevant cancer-related genetic and epigenetic modifications for discovering more detailed information on the tumor characterization. Although larger cohort trial with longitudinal analyses are needed, high prevalence of DDR gene and Tp53 mutation in our study instills promising hope for therapeutic interventions in this unique cohort.
The minimally invasive liquid biopsy shows a great promise as both diagnostic and prognostic tool in the personalized clinical management of advanced prostate, and NSCLC in the veteran patient population with unique demographic characteristics. De novo metastatic prostate cancer is more common in veterans when compared with the general population, and therefore veterans may benefit by liquid biopsy. Differences in the frequency of genetic mutations (DDR, TP53, AR) in this cohort provides valuable information for disease progression, lack of response, mechanism of resistance to the implemented therapy and clinical decision making. Precision oncology can be further tailored for this cohort by focusing on DNA repair genes and Tp53 mutations for future targeted therapy.
1
9
16. Institute of Medicine (US) Committee to Review the Health Effects in Vietnam Veterans of Exposure to Herbicides (Fourth Biennial Update). Veterans and Agent Orange: Update 2002. National Academies Press (US); 2003.
17. Eibner C, Krull H, Brown KM, et al. Current and projected characteristics and unique health care needs of the patient population served by the Department of Veterans Affairs. Rand Health Q. 2016;5(4):13. Published 2016 May 9.
18. Saarenheimo J, Eigeliene N, Andersen H, Tiirola M, Jekunen A. The value of liquid biopsies for guiding therapy decisions in non-small cell lung cancer. Front Oncol. 2019;9:129. Published 2019 Mar 5.doi:10.3389/fonc.2019.00129
19
20
21
22
23
24
25
26
27
28
29
30
31. Antonarakis ES, Lu C, Luber B, et al. Clinical significance of androgen receptor splice variant-7 mRNA detection in circulating tumor cells of men with metastatic castration-resistant prostate cancer treated with first- and second-line abiraterone and enzalutamide. J Clin Oncol. 2017;35(19):2149-2156. doi:10.1200/JCO.2016.70.1961
32

33. Jung A, Kirchner T. Liquid biopsy in tumor genetic diagnosis. Dtsch Arztebl Int. 2018;115(10):169-174. doi:10.3238/arztebl.2018.0169
34. Brennan S, Wang AR, Beyer H, et al. Androgen receptor as a potential target in non-small cell lung cancer. Cancer Res. 2017;77(Suppl13): abstract nr 4121. doi:10.1158/1538-7445.AM2017-4121
35. Wang AR, Beyer H, Brennan S, et al. Androgen receptor drives differential gene expression in KRAS-mediated non-small cell lung cancer. Cancer Res. 2018;78(Suppl 13): abstract nr 3946. doi:10.1158/1538-7445.AM2018-3946
The advent of liquid biopsies targeting genetic mutations in solid tumors is a major milestone in the field of precision oncology.1 Conventional methods of obtaining tissue for molecular studies are limited by sample size and often do not represent the entire bulk of the tumor.2 This newer minimally invasive, revolutionary technique analyzes circulating cell-free DNA carrying tumor-specific alterations (circulating tumor DNA [ctDNA]) in peripheral blood and detects signature genomic alterations.1 Tp53 mutations have been reported in 25 to 40% of prostatic cancers and > 50% of non-small cell lung cancers (NSCLC), being more common in late-stage and hormone refractory prostate cancers.3,4 Tp53 mutation has been found to be associated with poor prognosis and increased germline mutations.5
The veteran patient population has distinct demographic characteristics that make veterans more vulnerable to genetic mutations and malignancies, including risk of exposure to Agent Orange, smoking, substance abuse, and asbestos. This area is understudied and extremely sparse in the literature for frequency of genetic mutations, risk factors in solid malignancies occurring in the veteran patient population, and the clinical impact of these risk factors. We herein present a quality assurance study for the utility of liquid biopsies regarding the frequency of DNA damage repair (DDR) gene, Tp53, and androgen receptor (AR) mutations. The clinical impact in advanced lung and prostate cancers in the veteran patient population and frequency are the quality assurance observations that are the study endpoints.
Methods
We reviewed for quality assurance documentation from the Foundation Medicine (www.foundationmedicine.com) cancer biomarker tests on liquid biopsies performed at the Corporal Michael J. Crescenz Veteran Affairs Medical Center in Philadelphia, Pennsylvania from May 2019 to April 15, 2020. All biopsies were performed on cancers with biochemical, imaging or tissue evidence of advanced tumor progression. The testing was performed on advanced solid malignancies, including NSCLC, prostate adenocarcinoma, and metastatic colon cancer. Statistical data for adequacy; cases with notable mutations; frequency; and type of mutations of AR, DDR, and Tp53 were noted. General and specific risk factors associated with the veteran patient population were studied and matched with the type of mutations (Table 1).
Results
Thirty-one liquid biopsies were performed over this period—23 for prostate cancer, 7 for patients with lung cancer patients, and 1 for a patient with colon cancer. Of 31 cases, sensitivity/adequacy of liquid biopsy for genetic mutation was detected in 29 (93.5%) cases (Figure 1). Two inadequate biopsies (both from patients with prostate cancer) were excluded from the study, leaving 29 liquid biopsies with adequate ctDNA for analysis that were considered for further statistical purpose—21 prostate, 7 lung, and 1 colon cancer.
Multiple (common and different) genetic mutations were identified; however, our study subcategorized the mutations into the those that were related to prostate cancer, lung cancer, and some common mutations that occur in both cancers. Only the significant ones will be discussed in this review and equivocal result for AR is excluded from this study. Of the 21 prostate cancers, 4 (19.0%) had directed the targeted therapy to driver mutation (AR being most common in prostate cancer), while KRAS mutation, which was more common in lung cancer, was detected in 2/7 (28.6%) lung cancers. Mutations common to both cancer types were DDR gene mutations, which is a broad name for numerous genes including CDK12, ATM, and CHEK2.
Of all cases irrespective of the cancer type, 23/29 (79.3%) showed notable mutations. DDR gene mutations were found in 6 of 21 (28.5%) patients with prostate cancer and 8 of 23 (34.7%) patients with advanced prostate and lung cancers, indicating poor outcome and possible resistance to the current therapy. Of 23 patients showing mutations irrespective of the cancer type, 15 (65.2%) harbored Tp53 mutations, which is much more frequent in veteran patient population when compared with the literature. Fifteen of the 31 (48.4%) total patients were Vietnam War-era veterans who were potentially exposed to Agent Orange and 20 (64.5%) patients who were not Vietnam War-era veterans had a history that included smoking (Figure 2).
Discussion
The veteran patient population is a unique cohort due to its distinct demographic characteristics with a high volume of cancer cases diagnosed each year. According to data from VA Central Cancer Registry (VACCR), the most frequently diagnosed cancers are prostate (29%) and lung (18%).6
Liquid biopsy is a novel, promising technology that uses ctDNA and circulating tumor cells in peripheral blood for detecting genetic alterations through next generation sequencing.7-9 The advent of this minimally invasive, revolutionary technology has been a breakthrough in the field of precision oncology for prognosis, to monitor treatment response or resistance to therapy and further personalize cancer therapy.9,10
Comprehensive genomic profiling by liquid biopsy has many advantages over the molecular studies performed on tissue biopsy. Due to the tumor heterogeneity, tissue samples may not represent the full profile of the tumor genomics of cancer, while liquid biopsy has full presentation of the disease.11,12 Many times, tissue biopsy may be limited by a sample size that precludes full genetic profiling in addition to higher total cost, potential technical issues during processing, and possible side effects of the biopsy procedure.7,13 Additionally, as the tumor progresses, new driver mutations other than the ones previously detected on the primary tissue may emerge, which can confer resistance to the existing therapy.7,13
Advanced prostatic and lung carcinomas with biochemical, distant organ, or bony progression harbor unique signature genetic mutations indicating poor prognosis, lack of response or resistance to the existing therapy, and high risk of relapse.14,15 Some of the unique characteristics of the veteran patient population include a more aged patient population multiple comorbidities, higher frequency of > 1 type of cancer, advanced cancer stage at presentation, and specific risks factors such as exposure to Agent Orange in veterans who served during the Vietnam War era.16,17 We studied the utility of liquid biopsy in cancer care, including type and incidence of genomic alterations associated with advanced prostate and lung cancers, in this unique patient population.
The amount of cell-free DNA (cfDNA), also known as ctDNA varies widely in cancer patients. Some of the factors associated with low concentration of cfDNA are disease stage, intervening therapy, proliferation rates, and tumor vascularization.18,19 In the peripheral blood, of the total cfDNA, fractions of cfDNA varies from 0.01 to 90%.18,19 All samples containing ≥ 20 ng cfDNA (20 - 100 ng) were subjected to the hybrid capture-based NGS FoundationACT assay.20 In our study, 2 specimens did not meet the minimum criteria of adequacy (20 ng cfDNA); however, the overall adequacy rate for the detection of mutation, irrespective of the cancer type was 29 of 31 (93.5%) with only 2 inadequate samples. This rate is higher than the rate reported in the literature, which is about 70%.20
Significant differences were encountered in the incidence of DNA damage repair genes including Tp53 mutations when compared with those in the general patient population (Table 2). According to recent National Comprehensive Cancer Network (NCCN) guidelines, all prostate cancers should be screened for DDR gene mutations as these genes are common in aggressive prostate cancers and strongly associated with poor outcomes and shortened survival. Due to relatively high frequency of DDR gene mutations in advanced prostatic cancers, liquid biopsy in patients with these advanced stage prostate cancers may be a useful tool in clinical decision making and exploring targeted therapy.20
Mutations in BRCA2, ATM, CDK12, and CHEK2 (DDR gene family) are common. Incidence of ATM and CDK12 mutations in the literature is 3 to 6% of cases.21 Of 21 liquid biopsies of advanced prostate cancer patients, we found combined DDR gene mutation of ATM, CHEK2, and CDK12 genes in 6 (28.5%) cases, which is substantially higher than the 3 to 6% rate reported in the literature.21-24 Of the 23 patients who had notable mutations in our liquid biopsies, including both advanced prostate and lung cancer cases, 8 (34.7%) also showed mutation of the genes of DDR family. Our study did not show BRCA2 mutation, which is otherwise common in the literature.
We also evaluated the frequency of the most commonly occurring genetic mutations, Tp53 in advanced solid malignancies, especially advanced prostate and NSCLC. Previous studies have reported Tp53 mutation in association with risk factors (carcinogens) of cancer and have been a surrogate marker of poor survival or lack of response of therapy.25 Knowledge of Tp53 mutation is crucial for closer disease monitoring, preparing the patient for rapid progression, and encouraging the physician to prepare future lines of therapy.25-27 Although Tp53 mutation varies with histologic type and tissue of origin, Beltran and colleagues reported it in 30 to 40% of tumors, while Robles and colleagues reported about 40 to 42% incidence.25,27
Our study showed notable mutations in 23 of 29 adequate cases. Further, our study showed a high frequency of mutated Tp53 in 65.2% of combined advanced prostate and NSCLC cases. We then correlated cases of Vietnam War-era veterans with risk potential of Agent Orange exposure and Tp53 mutation. We found 7 of 15 Vietnam War-era veterans were positive for Tp53 mutations irrespective of the cancer type. The high incidence of Tp53 mutations in advanced prostate and lung carcinomas in the veteran patient population makes this tumor marker an aspiration not only as a surrogate of aggressive disease and tumor progression, but also as a key marker for targeted therapy in advanced prostate and lung cancers with loss of Tp53 function (Figure 3).
Mutations and amplifications in the AR gene are fundamental to progression of prostate cancer associated with advanced, hormone-refractory prostate cancer with the potential for targeted therapy with AR inhibitors. In our study, AR amplification was detected in 4 of 21 (19%) advanced prostate cancer cases, which is significantly lower than the 30 to 50% previously reported in the literature.28-32 Neither AR amplification or mutation was noted in advanced NSCLC in our study as previously reported in literature by Brennan and colleagues and Wang and colleagues.33-35 This is significant as it provides a pathway for future studies to focus on additional driver mutations for targeted therapies in advanced prostate carcinoma. To date, AR gene mutation does not play a role for personalized therapy in advanced NSCLC. Perhaps, a large cohort study with longitudinal analysis is needed for absolutely ruling out the possibility of personalized medicine in advanced lung cancer using this biomarker.
Conclusions
Liquid biopsy successfully provides precision-based oncology and information for decision making in this unique population of veterans. Difference in frequency of the genetic mutations in this cohort can provide future insight into disease progression, lack of response, and mechanism of resistance to the implemented therapy. Future studies focused on this veteran patient population are needed for developing targeted therapies and patient tailored oncologic therapy. ctDNA has a high potential for monitoring clinically relevant cancer-related genetic and epigenetic modifications for discovering more detailed information on the tumor characterization. Although larger cohort trial with longitudinal analyses are needed, high prevalence of DDR gene and Tp53 mutation in our study instills promising hope for therapeutic interventions in this unique cohort.
The minimally invasive liquid biopsy shows a great promise as both diagnostic and prognostic tool in the personalized clinical management of advanced prostate, and NSCLC in the veteran patient population with unique demographic characteristics. De novo metastatic prostate cancer is more common in veterans when compared with the general population, and therefore veterans may benefit by liquid biopsy. Differences in the frequency of genetic mutations (DDR, TP53, AR) in this cohort provides valuable information for disease progression, lack of response, mechanism of resistance to the implemented therapy and clinical decision making. Precision oncology can be further tailored for this cohort by focusing on DNA repair genes and Tp53 mutations for future targeted therapy.
The advent of liquid biopsies targeting genetic mutations in solid tumors is a major milestone in the field of precision oncology.1 Conventional methods of obtaining tissue for molecular studies are limited by sample size and often do not represent the entire bulk of the tumor.2 This newer minimally invasive, revolutionary technique analyzes circulating cell-free DNA carrying tumor-specific alterations (circulating tumor DNA [ctDNA]) in peripheral blood and detects signature genomic alterations.1 Tp53 mutations have been reported in 25 to 40% of prostatic cancers and > 50% of non-small cell lung cancers (NSCLC), being more common in late-stage and hormone refractory prostate cancers.3,4 Tp53 mutation has been found to be associated with poor prognosis and increased germline mutations.5
The veteran patient population has distinct demographic characteristics that make veterans more vulnerable to genetic mutations and malignancies, including risk of exposure to Agent Orange, smoking, substance abuse, and asbestos. This area is understudied and extremely sparse in the literature for frequency of genetic mutations, risk factors in solid malignancies occurring in the veteran patient population, and the clinical impact of these risk factors. We herein present a quality assurance study for the utility of liquid biopsies regarding the frequency of DNA damage repair (DDR) gene, Tp53, and androgen receptor (AR) mutations. The clinical impact in advanced lung and prostate cancers in the veteran patient population and frequency are the quality assurance observations that are the study endpoints.
Methods
We reviewed for quality assurance documentation from the Foundation Medicine (www.foundationmedicine.com) cancer biomarker tests on liquid biopsies performed at the Corporal Michael J. Crescenz Veteran Affairs Medical Center in Philadelphia, Pennsylvania from May 2019 to April 15, 2020. All biopsies were performed on cancers with biochemical, imaging or tissue evidence of advanced tumor progression. The testing was performed on advanced solid malignancies, including NSCLC, prostate adenocarcinoma, and metastatic colon cancer. Statistical data for adequacy; cases with notable mutations; frequency; and type of mutations of AR, DDR, and Tp53 were noted. General and specific risk factors associated with the veteran patient population were studied and matched with the type of mutations (Table 1).
Results
Thirty-one liquid biopsies were performed over this period—23 for prostate cancer, 7 for patients with lung cancer patients, and 1 for a patient with colon cancer. Of 31 cases, sensitivity/adequacy of liquid biopsy for genetic mutation was detected in 29 (93.5%) cases (Figure 1). Two inadequate biopsies (both from patients with prostate cancer) were excluded from the study, leaving 29 liquid biopsies with adequate ctDNA for analysis that were considered for further statistical purpose—21 prostate, 7 lung, and 1 colon cancer.
Multiple (common and different) genetic mutations were identified; however, our study subcategorized the mutations into the those that were related to prostate cancer, lung cancer, and some common mutations that occur in both cancers. Only the significant ones will be discussed in this review and equivocal result for AR is excluded from this study. Of the 21 prostate cancers, 4 (19.0%) had directed the targeted therapy to driver mutation (AR being most common in prostate cancer), while KRAS mutation, which was more common in lung cancer, was detected in 2/7 (28.6%) lung cancers. Mutations common to both cancer types were DDR gene mutations, which is a broad name for numerous genes including CDK12, ATM, and CHEK2.
Of all cases irrespective of the cancer type, 23/29 (79.3%) showed notable mutations. DDR gene mutations were found in 6 of 21 (28.5%) patients with prostate cancer and 8 of 23 (34.7%) patients with advanced prostate and lung cancers, indicating poor outcome and possible resistance to the current therapy. Of 23 patients showing mutations irrespective of the cancer type, 15 (65.2%) harbored Tp53 mutations, which is much more frequent in veteran patient population when compared with the literature. Fifteen of the 31 (48.4%) total patients were Vietnam War-era veterans who were potentially exposed to Agent Orange and 20 (64.5%) patients who were not Vietnam War-era veterans had a history that included smoking (Figure 2).
Discussion
The veteran patient population is a unique cohort due to its distinct demographic characteristics with a high volume of cancer cases diagnosed each year. According to data from VA Central Cancer Registry (VACCR), the most frequently diagnosed cancers are prostate (29%) and lung (18%).6
Liquid biopsy is a novel, promising technology that uses ctDNA and circulating tumor cells in peripheral blood for detecting genetic alterations through next generation sequencing.7-9 The advent of this minimally invasive, revolutionary technology has been a breakthrough in the field of precision oncology for prognosis, to monitor treatment response or resistance to therapy and further personalize cancer therapy.9,10
Comprehensive genomic profiling by liquid biopsy has many advantages over the molecular studies performed on tissue biopsy. Due to the tumor heterogeneity, tissue samples may not represent the full profile of the tumor genomics of cancer, while liquid biopsy has full presentation of the disease.11,12 Many times, tissue biopsy may be limited by a sample size that precludes full genetic profiling in addition to higher total cost, potential technical issues during processing, and possible side effects of the biopsy procedure.7,13 Additionally, as the tumor progresses, new driver mutations other than the ones previously detected on the primary tissue may emerge, which can confer resistance to the existing therapy.7,13
Advanced prostatic and lung carcinomas with biochemical, distant organ, or bony progression harbor unique signature genetic mutations indicating poor prognosis, lack of response or resistance to the existing therapy, and high risk of relapse.14,15 Some of the unique characteristics of the veteran patient population include a more aged patient population multiple comorbidities, higher frequency of > 1 type of cancer, advanced cancer stage at presentation, and specific risks factors such as exposure to Agent Orange in veterans who served during the Vietnam War era.16,17 We studied the utility of liquid biopsy in cancer care, including type and incidence of genomic alterations associated with advanced prostate and lung cancers, in this unique patient population.
The amount of cell-free DNA (cfDNA), also known as ctDNA varies widely in cancer patients. Some of the factors associated with low concentration of cfDNA are disease stage, intervening therapy, proliferation rates, and tumor vascularization.18,19 In the peripheral blood, of the total cfDNA, fractions of cfDNA varies from 0.01 to 90%.18,19 All samples containing ≥ 20 ng cfDNA (20 - 100 ng) were subjected to the hybrid capture-based NGS FoundationACT assay.20 In our study, 2 specimens did not meet the minimum criteria of adequacy (20 ng cfDNA); however, the overall adequacy rate for the detection of mutation, irrespective of the cancer type was 29 of 31 (93.5%) with only 2 inadequate samples. This rate is higher than the rate reported in the literature, which is about 70%.20
Significant differences were encountered in the incidence of DNA damage repair genes including Tp53 mutations when compared with those in the general patient population (Table 2). According to recent National Comprehensive Cancer Network (NCCN) guidelines, all prostate cancers should be screened for DDR gene mutations as these genes are common in aggressive prostate cancers and strongly associated with poor outcomes and shortened survival. Due to relatively high frequency of DDR gene mutations in advanced prostatic cancers, liquid biopsy in patients with these advanced stage prostate cancers may be a useful tool in clinical decision making and exploring targeted therapy.20
Mutations in BRCA2, ATM, CDK12, and CHEK2 (DDR gene family) are common. Incidence of ATM and CDK12 mutations in the literature is 3 to 6% of cases.21 Of 21 liquid biopsies of advanced prostate cancer patients, we found combined DDR gene mutation of ATM, CHEK2, and CDK12 genes in 6 (28.5%) cases, which is substantially higher than the 3 to 6% rate reported in the literature.21-24 Of the 23 patients who had notable mutations in our liquid biopsies, including both advanced prostate and lung cancer cases, 8 (34.7%) also showed mutation of the genes of DDR family. Our study did not show BRCA2 mutation, which is otherwise common in the literature.
We also evaluated the frequency of the most commonly occurring genetic mutations, Tp53 in advanced solid malignancies, especially advanced prostate and NSCLC. Previous studies have reported Tp53 mutation in association with risk factors (carcinogens) of cancer and have been a surrogate marker of poor survival or lack of response of therapy.25 Knowledge of Tp53 mutation is crucial for closer disease monitoring, preparing the patient for rapid progression, and encouraging the physician to prepare future lines of therapy.25-27 Although Tp53 mutation varies with histologic type and tissue of origin, Beltran and colleagues reported it in 30 to 40% of tumors, while Robles and colleagues reported about 40 to 42% incidence.25,27
Our study showed notable mutations in 23 of 29 adequate cases. Further, our study showed a high frequency of mutated Tp53 in 65.2% of combined advanced prostate and NSCLC cases. We then correlated cases of Vietnam War-era veterans with risk potential of Agent Orange exposure and Tp53 mutation. We found 7 of 15 Vietnam War-era veterans were positive for Tp53 mutations irrespective of the cancer type. The high incidence of Tp53 mutations in advanced prostate and lung carcinomas in the veteran patient population makes this tumor marker an aspiration not only as a surrogate of aggressive disease and tumor progression, but also as a key marker for targeted therapy in advanced prostate and lung cancers with loss of Tp53 function (Figure 3).
Mutations and amplifications in the AR gene are fundamental to progression of prostate cancer associated with advanced, hormone-refractory prostate cancer with the potential for targeted therapy with AR inhibitors. In our study, AR amplification was detected in 4 of 21 (19%) advanced prostate cancer cases, which is significantly lower than the 30 to 50% previously reported in the literature.28-32 Neither AR amplification or mutation was noted in advanced NSCLC in our study as previously reported in literature by Brennan and colleagues and Wang and colleagues.33-35 This is significant as it provides a pathway for future studies to focus on additional driver mutations for targeted therapies in advanced prostate carcinoma. To date, AR gene mutation does not play a role for personalized therapy in advanced NSCLC. Perhaps, a large cohort study with longitudinal analysis is needed for absolutely ruling out the possibility of personalized medicine in advanced lung cancer using this biomarker.
Conclusions
Liquid biopsy successfully provides precision-based oncology and information for decision making in this unique population of veterans. Difference in frequency of the genetic mutations in this cohort can provide future insight into disease progression, lack of response, and mechanism of resistance to the implemented therapy. Future studies focused on this veteran patient population are needed for developing targeted therapies and patient tailored oncologic therapy. ctDNA has a high potential for monitoring clinically relevant cancer-related genetic and epigenetic modifications for discovering more detailed information on the tumor characterization. Although larger cohort trial with longitudinal analyses are needed, high prevalence of DDR gene and Tp53 mutation in our study instills promising hope for therapeutic interventions in this unique cohort.
The minimally invasive liquid biopsy shows a great promise as both diagnostic and prognostic tool in the personalized clinical management of advanced prostate, and NSCLC in the veteran patient population with unique demographic characteristics. De novo metastatic prostate cancer is more common in veterans when compared with the general population, and therefore veterans may benefit by liquid biopsy. Differences in the frequency of genetic mutations (DDR, TP53, AR) in this cohort provides valuable information for disease progression, lack of response, mechanism of resistance to the implemented therapy and clinical decision making. Precision oncology can be further tailored for this cohort by focusing on DNA repair genes and Tp53 mutations for future targeted therapy.
1
9
16. Institute of Medicine (US) Committee to Review the Health Effects in Vietnam Veterans of Exposure to Herbicides (Fourth Biennial Update). Veterans and Agent Orange: Update 2002. National Academies Press (US); 2003.
17. Eibner C, Krull H, Brown KM, et al. Current and projected characteristics and unique health care needs of the patient population served by the Department of Veterans Affairs. Rand Health Q. 2016;5(4):13. Published 2016 May 9.
18. Saarenheimo J, Eigeliene N, Andersen H, Tiirola M, Jekunen A. The value of liquid biopsies for guiding therapy decisions in non-small cell lung cancer. Front Oncol. 2019;9:129. Published 2019 Mar 5.doi:10.3389/fonc.2019.00129
19
20
21
22
23
24
25
26
27
28
29
30
31. Antonarakis ES, Lu C, Luber B, et al. Clinical significance of androgen receptor splice variant-7 mRNA detection in circulating tumor cells of men with metastatic castration-resistant prostate cancer treated with first- and second-line abiraterone and enzalutamide. J Clin Oncol. 2017;35(19):2149-2156. doi:10.1200/JCO.2016.70.1961
32

33. Jung A, Kirchner T. Liquid biopsy in tumor genetic diagnosis. Dtsch Arztebl Int. 2018;115(10):169-174. doi:10.3238/arztebl.2018.0169
34. Brennan S, Wang AR, Beyer H, et al. Androgen receptor as a potential target in non-small cell lung cancer. Cancer Res. 2017;77(Suppl13): abstract nr 4121. doi:10.1158/1538-7445.AM2017-4121
35. Wang AR, Beyer H, Brennan S, et al. Androgen receptor drives differential gene expression in KRAS-mediated non-small cell lung cancer. Cancer Res. 2018;78(Suppl 13): abstract nr 3946. doi:10.1158/1538-7445.AM2018-3946
1
9
16. Institute of Medicine (US) Committee to Review the Health Effects in Vietnam Veterans of Exposure to Herbicides (Fourth Biennial Update). Veterans and Agent Orange: Update 2002. National Academies Press (US); 2003.
17. Eibner C, Krull H, Brown KM, et al. Current and projected characteristics and unique health care needs of the patient population served by the Department of Veterans Affairs. Rand Health Q. 2016;5(4):13. Published 2016 May 9.
18. Saarenheimo J, Eigeliene N, Andersen H, Tiirola M, Jekunen A. The value of liquid biopsies for guiding therapy decisions in non-small cell lung cancer. Front Oncol. 2019;9:129. Published 2019 Mar 5.doi:10.3389/fonc.2019.00129
19
20
21
22
23
24
25
26
27
28
29
30
31. Antonarakis ES, Lu C, Luber B, et al. Clinical significance of androgen receptor splice variant-7 mRNA detection in circulating tumor cells of men with metastatic castration-resistant prostate cancer treated with first- and second-line abiraterone and enzalutamide. J Clin Oncol. 2017;35(19):2149-2156. doi:10.1200/JCO.2016.70.1961
32

33. Jung A, Kirchner T. Liquid biopsy in tumor genetic diagnosis. Dtsch Arztebl Int. 2018;115(10):169-174. doi:10.3238/arztebl.2018.0169
34. Brennan S, Wang AR, Beyer H, et al. Androgen receptor as a potential target in non-small cell lung cancer. Cancer Res. 2017;77(Suppl13): abstract nr 4121. doi:10.1158/1538-7445.AM2017-4121
35. Wang AR, Beyer H, Brennan S, et al. Androgen receptor drives differential gene expression in KRAS-mediated non-small cell lung cancer. Cancer Res. 2018;78(Suppl 13): abstract nr 3946. doi:10.1158/1538-7445.AM2018-3946