User login
Diagnosis of Complicated Pneumonia
Community‐acquired pneumonia (CAP), the most common serious bacterial infection in childhood, may be complicated by parapneumonic effusion (ie, complicated pneumonia).1, 2 The clinical outcome of children with complicated pneumonia is directly influenced by the causative organism, and precise identification of the infectious agent has the potential to improve clinical management.35 The incidence of complicated pneumonia has recently increased, highlighting the need to better understand the reasons for this changing epidemiology.68 However, using conventional bacterial culture, bacteria are isolated from clinical samples obtained from children with complicated pneumonia in only 17%42% of cases.1, 9, 10 The low rate of positive culture results may be related to stringent bacterial growth requirements or to administration of broad‐spectrum antibiotics before obtaining blood or pleural fluid samples for culture.
Unlike bacterial culture, polymerase chain reaction (PCR) relies on the detection of bacterial DNA rather than on the recovery of viable bacteria and is therefore less affected by the prior administration of antibiotics. Broad‐range or universal primer 16S rRNA PCR detects a conserved region of the 16S ribosomal RNA (rRNA) gene and can detect a wide range of bacterial species with a single assay.11 However, few studies have evaluated the role of 16S rRNA PCR in detecting bacteria in the pleural fluid of children with complicated pneumonia.12, 13 The purpose of this study was to determine the frequency of positive blood and pleural fluid cultures in children with complicated pneumonia, and to determine whether broad‐range 16S rRNA PCR can increase the proportion of children with an identifiable microbiologic cause of complicated pneumonia.
Methods
Study Design, Setting, and Participants
This prospective cohort study was conducted at The Children's Hospital of Philadelphia (CHOP), an urban tertiary care children's hospital. The proposal was approved by the CHOP Committees for the Protection of Human Subjects. Patients were eligible for participation if they were 18 years of age, admitted to the hospital between October 1, 2007 and March 31, 2010, and diagnosed with complicated pneumonia. Parental informed consent was obtained for all patients, and verbal assent was obtained for all children over seven years of age. Patients with chronic medical conditions predisposing them to severe or recurrent pneumonia, such as human immunodeficiency virus, malignancy, and sickle cell disease, were excluded. The study team had no role in the clinical management of study patients. As this test is still considered experimental, pleural fluid 16S rRNA PCR was not performed until the patient was discharged from the hospital; these test results were not shared with the treating physicians.
Study Definition
Complicated pneumonia was defined by a temperature >38.0C, and the presence of lung parenchymal infiltrates and pleural effusions of any size or character on chest radiography or computed tomography.
Microbiologic MethodsConventional Culture
Pleural fluid or blood (4 mL) was inoculated onto a single pediatric BacT/Alert FAN bottle (BioMrieux, Durham, NC) which was immediately transported to the laboratory at room temperature. Additional pleural fluid was also submitted to the laboratory for a cytospin Gram stain. Once received in the laboratory, the bottles were immediately loaded into the BacT/Alert instrument. Bottles were automatically checked by the instrument for production of CO2 every ten minutes, and remained in the instrument for a total of five days. Bottles flagged as positive by the BacT/Alert system were removed from the instrument, subcultured to agar plates, and Gram stained. Bacterial isolates were identified and the antibiotic susceptibility tested by conventional methods following Clinical and Laboratory Standards Institute guidelines.
Microbiological Methods16S rRNA PCR
The 16S rRNA primers and probe used in this study have been validated previously and have been used extensively for the identification of organisms from the Domain Bacteria.14 Escherichia coli (ATCC 25922) and Staphylococcus aureus (ATCC 29213) were used as Gram‐negative and Gram‐positive controls, respectively. DNA extraction was performed on a 300 l aliquot of pleural fluid using the MagMAX Total Nucleic Acid Isolation Kit (Applied Biosystems, Foster City, CA). Samples were disrupted using a Vortex adapter for 15 minutes at the highest setting. Tubes were then centrifuged at 16,000 relative centrifugal force (RCF) for three minutes, and DNA extraction was performed using the KingFisher Flex Automated Purification System (Thermo Scientific, Waltham, MA). All DNA extracts were frozen at 20C prior to use. Details of the 16S rRNA PCR assay optimization and validation appear in the Appendix.
DNA was amplified on an Applied Biosystems 7500 thermal cycler (ABI, Foster City, CA) using the primers, 16S F: 5‐GCT GTC ATC TCT TGT GGG CTG T‐3, 16S R: 5‐AAA CTC ATG GGA GCT GCT GGT T‐3, and the fluorescent labeled TaqMan probe, 16s Probe: 5‐Cy5/CCT GTC ATG CCC ACA CAA ATC TCT CC/BHQ‐2‐3. Human DNA (0.5 nanograms) was added to each assay as a standardized internal amplification control. The primers, AlbICF: 5‐GCT GTC ATC TCT TGT GGG CTG T‐3, AlbICR: 5‐AAA CTC ATG GGA GCT GCT GGT T‐3, and the TaqMan probe, AlbICP: 5‐Cy5/CCT GTC ATG CCC ACA CAA ATC TCT CC/BHQ‐2‐3 detect a region of the human albumin gene. Amplification was performed as follows: initial denaturation of 94C for 20 seconds, followed by 35 cycles of 94C for 10 seconds, 61C for 31 seconds, and 72C for 5 seconds. Each run included positive (E. coli ATCC 25922 DNA) and negative (nuclease free water) controls. A cycle threshold value of 30 or less indicated a positive sample. All positive samples were confirmed using the MicroSeq 500 16S rDNA Bacterial Identification kit (Applied Biosystems).
DNA Sequencing
DNA sequencing was performed directly on PCR products using an ABI 3730 with the BigDye Taq FS Terminator V3.1. Sequencing was completed at the University of Pennsylvania DNA Sequencing Facility, and sequence identification was performed by BLAST at the National Center for Biotechnology Information web site (
Statistical Analysis
Data were analyzed using STATA 10.0 (Stata Corp., College Station, TX). Categorical variables were described using frequencies and percentages, and were compared using the chi‐square test. Continuous variables were described using mean, median, intraquartile range (IQR), and range values. Binomial exact 95% confidence intervals (CIs) were calculated for the proportion of positive blood and pleural fluid cultures and pleural fluid 16S rRNA PCR tests.
Results
During the study period, 124 patients with complicated pneumonia were identified; the median age was 4.9 years (IQR, 2.78.8 years) and the ratio of female to male patients was 1:1. The racial distribution was as follows: non‐Hispanic white, 62 (50%); non‐Hispanic black, 43 (34%); and Hispanic, 7 (6%). The median length of stay was 11 days (range, 240 days; IQR, 811 days). Forty‐one (33.1%) patients were admitted to the intensive care unit, and nine (7.3%) patients required mechanical ventilation. Overall, 5 of the 71 blood cultures performed (7.0%; 95% CI: 2.3%15.7%) were positive.
Pleural fluid drainage was performed in 64 (51.6%) patients. The median duration of antibiotic treatment before pleural fluid drainage was two days (IQR, 04 days). Differences in duration of antibiotic pretreatment between those with and without pleural fluid drainage were not significant. Patients received beta‐lactam agents (76.6%), clindamycin (51.6%), and vancomycin (23.4%) before pleural drainage. Blood cultures were performed in 27 (45.0%) of 60 patients not undergoing pleural fluid drainage; 2 of these 27 (7.4%; 95% CI: 0.9%24.3%) blood cultures were positive, and Streptococcus pneumoniae was isolated in both cases.
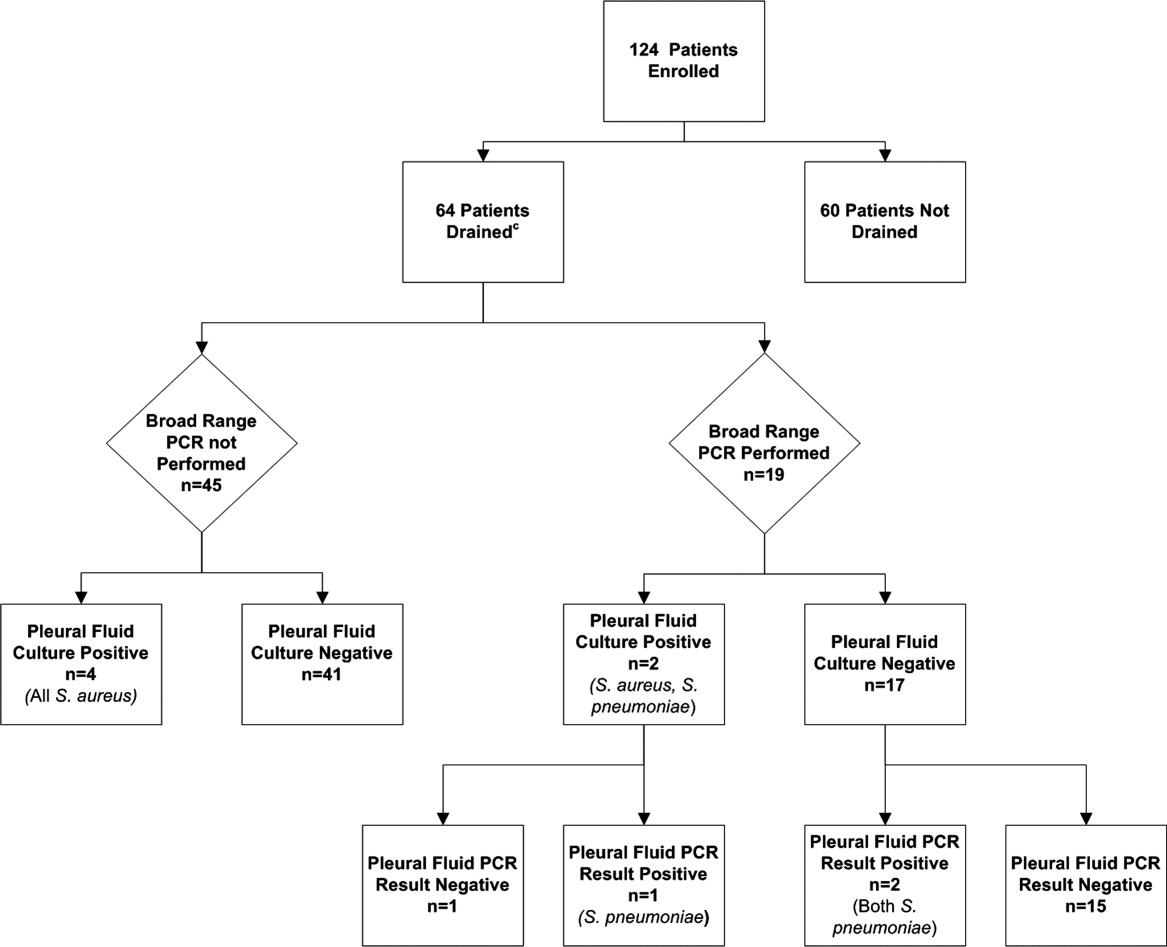
Using a combination of pleural fluid culture, blood culture, and broad‐range PCR, a microbiologic cause of infection was identified in 11 of 64 (17.2%; 95% CI: 8.9%28.7%) patients undergoing pleural fluid drainage (Figure 1). Patients with an identified bacterial cause were younger than those without an identified bacterial cause (Table 1). However, there were no statistically significant differences between the groups with and without identified bacteria with respect to sex, race, clinical examination findings and laboratory results at admission, duration of antibiotic therapy before pleural fluid drainage, or length of stay (Table 1). The median pleural fluid white blood cell count was 6880 per mm3 (IQR, 154227,918 per mm3).
| Causative Bacteria Identified | |||
|---|---|---|---|
| Demographics | Yes (n = 11) | No (n = 53) | P Value |
| |||
| Female sex | 3 (27.3%) | 29 (54.7%) | 0.10 |
| Age (years) | 2 (1.34.50) | 4.3 (3.1 8.1) | 0.03 |
| Race | 0.09 | ||
| Non‐Hispanic white | 3 (27.3%) | 29 (54.7%) | |
| Non‐Hispanic black | 7 (63.6%) | 13 (24.5%) | |
| Hispanic | 0 (0.0%) | 3 (5.6%) | |
| Other | 1 (9.1%) | 8 (15.1%) | |
| History of asthma | 2 (18.2%%) | 12 (22.6%) | 0.75 |
| Duration of antibiotics before drainage (days) | 0.5 (03) | 2 (04) | 0.43 |
| Intensive care unit admission | 7 (63.6%) | 25 (47.2%) | 0.32 |
| Initial temperature (C) | 37.8 (37.239.5) | 38.1 (37.639.3) | 0.76 |
| Initial oxygen saturation (%) | 94 (9397) | 95 (9396) | 0.97 |
| Peripheral white blood cell count (per mm3) | 21.4 (14.028.4) | 16.3 (10.922.6) | 0.12 |
| Platelet count (per mm3) | 434 (298546) | 402 (312575) | 0.95 |
Blood was obtained for culture from 44 (68.8%) of 64 patients that underwent pleural fluid drainage. Blood cultures were positive in three (6.8%; 95% CI: 1.4%18.7%) of these patients; causative bacteria were S. pneumoniae (n = 1), Haemophilus influenzae (n = 1), and Staphylococcus aureus (n = 1). Pleural fluid cultures were positive from 6 of 64 patients (9.3%; 95% CI: 3.5%19.3%) that underwent pleural fluid drainage; causative bacteria were Staphylococcus aureus (n = 5) and Streptococcus pneumoniae (n = 1). Three of the 19 pleural fluid samples (15.8%, CI 3.4%39.6%) tested by 16S rRNA PCR yielded positive results; S. pneumoniae was identified by DNA sequencing in all three of these samples (Figure 1). Of these three specimens, two had both negative Gram stains and negative pleural fluid cultures, while one specimen had Gram‐positive cocci in pairs identified on Gram stain along with a pleural fluid culture‐positive for S. pneumoniae. Of the 16 patients with negative PCR results, one was found to have a pleural fluid culture‐positive for S. aureus; no bacteria were detected on the pleural fluid Gram stain (Figure 1). Differences in the demographic characteristics between patients in whom broad‐range PCR was performed and not performed were not statistically significant (Table 2). Differences in the median length of hospital stay between patients who had PCR performed (9 days; IQR, 712 days) and those who did not (11 days; IQR, 914 days) were not statistically significant (P = 0.19). Antibiotic therapy was simplified from treatment with multiple antibiotics to a single antibiotic in each instance of a positive culture, though the number of positive cultures was too small to lead to meaningful reductions in antibiotic use or spectrum.
| PCR Performed | |||
|---|---|---|---|
| Demographics | Yes (N = 19) | No (N = 45) | P Value |
| |||
| Female sex | 9 (47.4) | 23 (51.1) | 0.78 |
| Age (years) | 4.9 (2.78.8) | 4.0 (2.88.1) | 0.76 |
| Race | 0.42 | ||
| Non‐Hispanic white | 10 (52.6%) | 22 (48.9%) | |
| Non‐Hispanic black | 5 (26.3%) | 15 (33.3%) | |
| Hispanic | 0 (0.0%) | 3 (6.7%) | |
| Other | 4 (21.1%) | 5 (11.1%) | |
| History of asthma | 5 (26.32%) | 9 (20.0%) | 0.58 |
| Duration of antibiotics before drainage (days) | 2 (03) | 1.5 (04) | 0.65 |
| Intensive care unit admission | 8 (42.1%) | 24 (53.3%) | 0.41 |
| Initial temperature (C) | 38.1 (37.339.4) | 38.0 (37.639.4) | 0.86 |
| Initial pulse oximetry (%) | 96 (9497) | 94 (9296) | 0.34 |
| Initial white blood cell count (1000 per mm3) | 16.1 (9.527.4) | 17.8 (12.622.8) | 0.93 |
| Platelet count (1000 per mm3) | 397 (312575) | 431 (303597) | 0.73 |
Discussion
Identification of the causative organism can improve the treatment of children with complicated pneumonia by enabling clinicians to target the infection with effective, narrow‐spectrum antibiotics. However, broad‐spectrum antibiotics are typically given before blood or pleural fluid samples are obtained, lowering the yield of conventional bacterial cultures. In our study, pleural fluid and blood cultures were infrequently positive. However, the use of 16S rRNA broad‐range PCR and DNA sequencing as ancillary tests only modestly improved our diagnostic yield.
Two prior studies have shown that broad‐range PCR analysis of pleural fluid can detect pathogenic organisms from pleural fluid even after the administration of antibiotics. Saglani et al.12 used bacterial culture and broad‐range PCR to analyze pleural fluid from 32 children with complicated pneumonia. Although the cohort had received a median of eight days of antibiotic therapy prior to fluid aspiration, a combination of broad‐range PCR and DNA sequencing identified organisms, predominantly S. pneumoniae, in 17 of 26 (65.4%) pleural fluid samples with negative culture. Five of the six culture‐positive samples also had positive PCR results, suggesting that a low proportion of the PCR results were false‐negatives. Le Monnier et al.13 cultured the pleural fluid from 78 children with complicated pneumonia; 15 samples were excluded from PCR testing because they grew bacteria other than S. pneumoniae. Broad‐range 16S rRNA PCR detected bacteria in 22 of 40 (55.0%) samples that were culture‐negative; subsequent DNA sequencing identified S. pneumoniae (n = 17), S. pyogenes (n = 3), S. aureus (n = 1), and H. influenzae (n = 1). S. pneumoniae was identified by 16S rRNA PCR followed by DNA sequencing in 20 of the 23 (87.0%) pleural fluid samples that grew S. pneumoniae on culture.
PCR with organism‐specific primers has also been shown to detect pathogenic bacteria in 35%70% of pleural fluid samples from patients with complicated pneumonia.15, 16 These rates of detection are comparable to the results of prior studies that used broad‐range PCR, and suggest that 16S rRNA and organism‐specific PCR are similarly sensitive tests.
Our study found that the proportion of positive results with broad‐range PCR was greater than the proportion observed with conventional pleural fluid culture. We were also able to identify S. pneumoniae by broad‐range PCR in two culture‐negative pleural fluid samples. The detection of S. pneumoniae by PCR in the setting of negative pleural fluid culture is not surprising, as most patients received empiric antibiotic therapy against S. pneumoniae before undergoing pleural fluid drainage. This prior antibiotic therapy would be expected to decrease the yield of pleural fluid culture but have a less significant impact on PCR. The reason for the failure to detect S. aureus by PCR from our clinical samples is not known. We explored whether this issue could be attributable to test characteristics during validation of our assay (as described in the technical Appendix), and S. aureus was consistently identified in pleural fluid samples spiked with S. aureus. While our diagnostic yield was low compared with prior studies that used broad‐range PCR, the distribution of causative bacteria was similar to prior studies of complicated pneumonia.9, 10 There were too few positive cultures among the specimens available for broad‐range PCR testing for us to reliably assess 16S rRNA PCR sensitivity and specificity. Thus, it was not possible to reliably assess the value of 16S rRNA PCR as an ancillary test in culture‐negative complicated pneumonia.
Several factors may have contributed to the low yield of broad‐range PCR in our study. First, all patients in our study received broad‐spectrum antibiotics prior to pleural fluid drainage; most patients received treatment with a beta‐lactam agent in combination with either clindamycin or vancomycin. Although PCR is less affected than culture by the prior administration of antibiotics, it is still possible that exposure to antibiotics accelerated degradation of the bacterial genome, thus decreasing the sensitivity of broad‐range PCR. The median duration of antibiotic therapy prior to drainage was shorter for patients in whom a bacterial pathogen was identified, compared with those in whom a pathogen was not identified. Though this difference was not statistically significant, this difference emphasizes that early pleural fluid collection may improve bacterial detection. The median duration of prior antibiotic therapy was eight days, in the study by Saglani et al.,12 however details regarding anti‐staphylococcal therapy were not reported. Second, several studies using 16S rRNA PCR in blood or cerebrospinal fluid (CSF) specimens for children with sepsis or meningitis, respectively, have noted decreased sensitivity for detection of Gram‐positive bacteria.1720 The cell wall composition makes Gram‐positive bacteria particularly difficult to lyse for DNA extraction. Prior studies used a different approach to DNA extraction compared with our study. Saglani et al.12 used the QIAmp minikit for DNA extraction (Qiagen Ltd, West Sussex, UK) and Le Monnier et al.13 used the MagnaPure System (Roche Diagnostics, Indianapolis, IN) for DNA extraction; additionally Saglani et al.12 included an additional 15 minutes of incubation at 95C following Proteinase K digestion to ensure complete lysis of the bacterial cells. We used the MagMAX Total Nucleic Acid Isolation Kit with a bead beating step to degrade the samples prior to lysis. It is unlikely that any of these differences contributed to differences in our study results. Third, human DNA may cause a nonspecific background signal, which could decrease PCR sensitivity. This issue may be particularly important in complicated pneumonia where the pleural fluid white blood cell counts are substantially elevated. Human DNA from white blood cells might be present at much higher concentrations than bacterial DNA, which would create a competitive advantage for binding of human, rather than bacterial, DNA on to the bead matrix.21 Saglani et al.12 did not report pleural fluid white blood cell counts for comparison. However, Le Monnier et al.13 reported pleural fluid white blood cell counts slightly higher than those observed in our patients. Thus, differences in pleural fluid white blood cell counts do not necessarily explain the differences in yield across studies. Fourth, while the universal primers used in our study have been validated, including for detection of S. aureus, in previous studies,14 Saglani et al.12 and Le Monnier et al.13 each used a different set of primers. It is unclear whether or not the choice of different primers affected the microbiologic yield.
An important limitation of this study is the relatively small number of patients with pleural fluid available for PCR testing combined with the low PCR yield. This limitation may have caused us to underestimate the true benefit of 16S rRNA PCR as an ancillary diagnostic test, as suggested by the wide confidence intervals around the estimates of PCR yield. Additionally, not all patients underwent pleural drainage, and not all patients undergoing pleural drainage had pleural fluid available for PCR testing. The latter issue would likely have only minimal impact on our study results, as there were no differences in demographics or clinical or laboratory features among those with and without pleural fluid available for PCR testing. However, differences among physicians, and across institutions, may play an important role in the decision to perform pleural drainage. We do not know whether patients undergoing pleural drainage at our institution were more or less likely to have bacteria detected by PCR than patients not undergoing drainage. If they were more likely to have bacteria detected, then our study would underestimate the benefit of PCR as an adjunct diagnostic test. However, there was no difference in the duration of antibiotic pretreatment between these two groups, and the yield from blood culture was similar.
In conclusion, blood and pleural fluid cultures infrequently identify the causative bacteria in children with complicated pneumonia. The use of broad‐range PCR increased the microbiologic yield only modestly. Further refinements to improve the diagnostic accuracy of broad‐range PCR testing are needed before this technique can be recommended for widespread use in clinical practice.
Appendix
Optimization and Validation of The 16S Assay
The assay was optimized for use with pleural fluid samples as follows. E. coli (ATCC 25922) and S. aureus (ATCC 29213) were grown overnight in 100 ml Brain Heart Infusion (BHI) broth. A suspension equivalent to a 0.5 MacFarland standard was prepared and was taken to represent approximately 1 108 colony‐forming units/mL. To prepare standard PCR curves, 1 in 10 dilutions were prepared in PBS (pH, 7.4) and DNA was extracted, in triplicate, using two different DNA extraction methods: the MagMAX Total Nucleic Acid Isolation Kit and the PrepMan Ultra Sample Preparation reagent system (Applied Biosystems, Foster City, CA), as described by the manufacturer. For the MagMax extraction, a 300 microliter sample was added to 200 microliters of lysis/binding solution in a bead tube. Samples were disrupted using a vortex adapter for 15 minutes at the highest setting. Tubes were then centrifuged at 16,000 relative centrifugal force (RCF) for three minutes. DNA extraction was performed using the KingFisher Flex Automated Purification System (Thermo Scientific). For the PrepMan Ultra extraction, a 300‐l sample was centrifuged for three minutes at 16,000 RCF. The supernatant fluid was discarded and the pellet was resuspended in 100 microliters of PrepMan Ultra Lysis Buffer and incubated at 100C for ten minutes. The sample was centrifuged for three minutes at 16,000 RCF, and a 10‐microliter aliquot of supernatant fluid was added to 90 microliters of nuclease free water. All DNA extracts were frozen at 20C prior to use. Plate counts, performed from the bacterial suspensions in triplicate, revealed approximately 1.3 108 colony‐forming units/mL in the original suspensions.
Pleural fluid samples were spiked with known concentrations (1, 3, 10, 30, 100, 300, and 1000 colony‐forming units/mL) of E. coli and S. aureus, and each sample was extracted five times with both DNA extraction systems. Each DNA extract was amplified on an Applied Biosystems 7500 thermal cycler (ABI) and a SmartCycler (Cepheid, Sunnyvale, CA). Analysis of the results obtained from the preliminary experiments indicated that the following combination of DNA extraction and PCR was optimal, and these conditions were used for all patient samples.
DNA was extracted from a 300‐microliter aliquot of pleural fluid using the MagMAX Total Nucleic Acid Isolation Kit (Ambion, Applied Biosystems) as described by the manufacturer. PCR was performed on an Applied Biosystems 7500 thermal cycler (ABI) using the primers, 16S F: 5‐GCT GTC ATC TCT TGT GGG CTG T‐3, 16S R: 5‐AAA CTC ATG GGA GCT GCT GGT T‐3, and the fluorescent labeled TaqMan probe, 16s Probe: 5‐Cy5/CCT GTC ATG CCC ACA CAA ATC TCT CC/BHQ‐2‐3. Human DNA (0.5 nanograms) was added to each assay to ensure that an adequate amount of human DNA was available to act as an internal amplification control. The primers, AlbICF: 5‐GCT GTC ATC TCT TGT GGG CTG T‐3, AlbICR: 5‐AAA CTC ATG GGA GCT GCT GGT T‐3, and the TaqMan probe, AlbICP: 5‐Cy5/CCT GTC ATG CCC ACA CAA ATC TCT CC/BHQ‐2‐3 detect a region of the human albumin gene. Amplification was performed as follows: initial denaturation of 94C for 20 seconds, followed by 35 cycles of 94C for 10 seconds, 61C for 31 seconds, and 72C for 5 seconds. Each run included positive (E. coli ATCC 25922 DNA) and negative (nuclease free water) controls. A cycle threshold value of 30 or less indicated a positive sample. All positive samples were confirmed using the MicroSeq 500 16S rDNA Bacterial Identification kit (Applied Biosystems).
- ,,, et al.An epidemiological investigation of a sustained high rate of pediatric parapneumonic empyema: risk factors and microbiological associations.Clin Infect Dis.2002;34:434–440.
- ,,,,.Primary early thoracoscopy and reduction in length of hospital stay and additional procedures among children with complicated pneumonia: results of a multicenter retrospective cohort study.Arch Pediatr Adolesc Med.2008;162:675–681.
- ,,, et al.Pulmonary manifestations in children with invasive community‐acquired Staphylococcus aureus infection.Clin Infect Dis.2005;41:583–590.
- ,,, et al.Association between Staphylococcus aureus strains carrying gene for Panton‐Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients.Lancet.2002;359:753–759.
- ,,, et al.Severe community‐onset pneumonia in healthy adults caused by methicillin‐resistant Staphylococcus aureus carrying the Panton‐Valentine leukocidin genes.Clin Infect Dis.2005;40:100–107.
- ,.Empyema hospitalizations increased in US children despite pneumococcal conjugate vaccine.Pediatrics.2010;125:26–33.
- ,,,.Increasing incidence of empyema complicating childhood community‐acquired pneumonia in the United States.Clin Infect Dis.2010;50:805–813.
- ,,,,.National hospitalization trends for pediatric pneumonia and associated complications.Pediatrics.2010;126:204–213.
- ,,, et al.Thoracoscopic decortication vs tube thoracostomy with fibrinolysis for empyema in children: a prospective, randomized trial.J Pediatr Surg.2009;44:106–111.
- ,,, et al.Comparison of urokinase and video‐assisted thoracoscopic surgery for treatment of childhood empyema.Am J Respir Crit Care Med.2006;174:221–227.
- ,.Development of broad‐range 16S rDNA PCR for use in the routine diagnostic clinical microbiology service.J Med Microbiol.2003;52:685–691.
- ,,,.Empyema: the use of broad range 16S rDNA PCR for pathogen detection.Arch Dis Child.2005;90:70–73.
- ,,, et al.Microbiological diagnosis of empyema in children: comparative evaluations by culture, polymerase chain reaction, and pneumococcal antigen detection in pleural fluids.Clin Infect Dis.2006;42:1135–1140.
- ,,,.Determination of bacterial load by real‐time PCR using a broad‐range (universal) probe and primers set.Microbiology.2002;148:257–266.
- ,,,,,.Pneumolysin polymerase chain reaction for diagnosis of pneumococcal pneumonia and empyema in children.Eur J Clin Microbiol Infect Dis.2006;25:783–789.
- ,,, et al.Pleural fluid PCR method for detection of Staphylococcus aureus, Streptococcus pneumoniae and Haemophilus influenzae in pediatric parapneumonic effusions.Respiration.2008;75:437–442.
- ,.16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls.J Clin Microbiol.2007;45:2761–2764.
- ,,,,.Evaluating the near‐term infant for early onset sepsis: progress and challenges to consider with 16S rDNA polymerase chain reaction testing.J Mol Diagn.2006;8:357–363.
- ,,,,.Preanalytic removal of human DNA eliminates false signals in general 16S rDNA PCR monitoring of bacterial pathogens in blood.Comp Immunol Microbiol Infect Dis.2009;32:207–219.
- ,,,,.Use of broad range16S rDNA PCR in clinical microbiology.J Microbiol Methods.2009;76:217–225.
- ,.Real‐time polymerase chain reaction for detecting bacterial DNA directly from blood of neonates being evaluated for sepsis.J Mol Diagn.2005;7:575–581.
Community‐acquired pneumonia (CAP), the most common serious bacterial infection in childhood, may be complicated by parapneumonic effusion (ie, complicated pneumonia).1, 2 The clinical outcome of children with complicated pneumonia is directly influenced by the causative organism, and precise identification of the infectious agent has the potential to improve clinical management.35 The incidence of complicated pneumonia has recently increased, highlighting the need to better understand the reasons for this changing epidemiology.68 However, using conventional bacterial culture, bacteria are isolated from clinical samples obtained from children with complicated pneumonia in only 17%42% of cases.1, 9, 10 The low rate of positive culture results may be related to stringent bacterial growth requirements or to administration of broad‐spectrum antibiotics before obtaining blood or pleural fluid samples for culture.
Unlike bacterial culture, polymerase chain reaction (PCR) relies on the detection of bacterial DNA rather than on the recovery of viable bacteria and is therefore less affected by the prior administration of antibiotics. Broad‐range or universal primer 16S rRNA PCR detects a conserved region of the 16S ribosomal RNA (rRNA) gene and can detect a wide range of bacterial species with a single assay.11 However, few studies have evaluated the role of 16S rRNA PCR in detecting bacteria in the pleural fluid of children with complicated pneumonia.12, 13 The purpose of this study was to determine the frequency of positive blood and pleural fluid cultures in children with complicated pneumonia, and to determine whether broad‐range 16S rRNA PCR can increase the proportion of children with an identifiable microbiologic cause of complicated pneumonia.
Methods
Study Design, Setting, and Participants
This prospective cohort study was conducted at The Children's Hospital of Philadelphia (CHOP), an urban tertiary care children's hospital. The proposal was approved by the CHOP Committees for the Protection of Human Subjects. Patients were eligible for participation if they were 18 years of age, admitted to the hospital between October 1, 2007 and March 31, 2010, and diagnosed with complicated pneumonia. Parental informed consent was obtained for all patients, and verbal assent was obtained for all children over seven years of age. Patients with chronic medical conditions predisposing them to severe or recurrent pneumonia, such as human immunodeficiency virus, malignancy, and sickle cell disease, were excluded. The study team had no role in the clinical management of study patients. As this test is still considered experimental, pleural fluid 16S rRNA PCR was not performed until the patient was discharged from the hospital; these test results were not shared with the treating physicians.
Study Definition
Complicated pneumonia was defined by a temperature >38.0C, and the presence of lung parenchymal infiltrates and pleural effusions of any size or character on chest radiography or computed tomography.
Microbiologic MethodsConventional Culture
Pleural fluid or blood (4 mL) was inoculated onto a single pediatric BacT/Alert FAN bottle (BioMrieux, Durham, NC) which was immediately transported to the laboratory at room temperature. Additional pleural fluid was also submitted to the laboratory for a cytospin Gram stain. Once received in the laboratory, the bottles were immediately loaded into the BacT/Alert instrument. Bottles were automatically checked by the instrument for production of CO2 every ten minutes, and remained in the instrument for a total of five days. Bottles flagged as positive by the BacT/Alert system were removed from the instrument, subcultured to agar plates, and Gram stained. Bacterial isolates were identified and the antibiotic susceptibility tested by conventional methods following Clinical and Laboratory Standards Institute guidelines.
Microbiological Methods16S rRNA PCR
The 16S rRNA primers and probe used in this study have been validated previously and have been used extensively for the identification of organisms from the Domain Bacteria.14 Escherichia coli (ATCC 25922) and Staphylococcus aureus (ATCC 29213) were used as Gram‐negative and Gram‐positive controls, respectively. DNA extraction was performed on a 300 l aliquot of pleural fluid using the MagMAX Total Nucleic Acid Isolation Kit (Applied Biosystems, Foster City, CA). Samples were disrupted using a Vortex adapter for 15 minutes at the highest setting. Tubes were then centrifuged at 16,000 relative centrifugal force (RCF) for three minutes, and DNA extraction was performed using the KingFisher Flex Automated Purification System (Thermo Scientific, Waltham, MA). All DNA extracts were frozen at 20C prior to use. Details of the 16S rRNA PCR assay optimization and validation appear in the Appendix.
DNA was amplified on an Applied Biosystems 7500 thermal cycler (ABI, Foster City, CA) using the primers, 16S F: 5‐GCT GTC ATC TCT TGT GGG CTG T‐3, 16S R: 5‐AAA CTC ATG GGA GCT GCT GGT T‐3, and the fluorescent labeled TaqMan probe, 16s Probe: 5‐Cy5/CCT GTC ATG CCC ACA CAA ATC TCT CC/BHQ‐2‐3. Human DNA (0.5 nanograms) was added to each assay as a standardized internal amplification control. The primers, AlbICF: 5‐GCT GTC ATC TCT TGT GGG CTG T‐3, AlbICR: 5‐AAA CTC ATG GGA GCT GCT GGT T‐3, and the TaqMan probe, AlbICP: 5‐Cy5/CCT GTC ATG CCC ACA CAA ATC TCT CC/BHQ‐2‐3 detect a region of the human albumin gene. Amplification was performed as follows: initial denaturation of 94C for 20 seconds, followed by 35 cycles of 94C for 10 seconds, 61C for 31 seconds, and 72C for 5 seconds. Each run included positive (E. coli ATCC 25922 DNA) and negative (nuclease free water) controls. A cycle threshold value of 30 or less indicated a positive sample. All positive samples were confirmed using the MicroSeq 500 16S rDNA Bacterial Identification kit (Applied Biosystems).
DNA Sequencing
DNA sequencing was performed directly on PCR products using an ABI 3730 with the BigDye Taq FS Terminator V3.1. Sequencing was completed at the University of Pennsylvania DNA Sequencing Facility, and sequence identification was performed by BLAST at the National Center for Biotechnology Information web site (
Statistical Analysis
Data were analyzed using STATA 10.0 (Stata Corp., College Station, TX). Categorical variables were described using frequencies and percentages, and were compared using the chi‐square test. Continuous variables were described using mean, median, intraquartile range (IQR), and range values. Binomial exact 95% confidence intervals (CIs) were calculated for the proportion of positive blood and pleural fluid cultures and pleural fluid 16S rRNA PCR tests.
Results
During the study period, 124 patients with complicated pneumonia were identified; the median age was 4.9 years (IQR, 2.78.8 years) and the ratio of female to male patients was 1:1. The racial distribution was as follows: non‐Hispanic white, 62 (50%); non‐Hispanic black, 43 (34%); and Hispanic, 7 (6%). The median length of stay was 11 days (range, 240 days; IQR, 811 days). Forty‐one (33.1%) patients were admitted to the intensive care unit, and nine (7.3%) patients required mechanical ventilation. Overall, 5 of the 71 blood cultures performed (7.0%; 95% CI: 2.3%15.7%) were positive.
Pleural fluid drainage was performed in 64 (51.6%) patients. The median duration of antibiotic treatment before pleural fluid drainage was two days (IQR, 04 days). Differences in duration of antibiotic pretreatment between those with and without pleural fluid drainage were not significant. Patients received beta‐lactam agents (76.6%), clindamycin (51.6%), and vancomycin (23.4%) before pleural drainage. Blood cultures were performed in 27 (45.0%) of 60 patients not undergoing pleural fluid drainage; 2 of these 27 (7.4%; 95% CI: 0.9%24.3%) blood cultures were positive, and Streptococcus pneumoniae was isolated in both cases.
Using a combination of pleural fluid culture, blood culture, and broad‐range PCR, a microbiologic cause of infection was identified in 11 of 64 (17.2%; 95% CI: 8.9%28.7%) patients undergoing pleural fluid drainage (Figure 1). Patients with an identified bacterial cause were younger than those without an identified bacterial cause (Table 1). However, there were no statistically significant differences between the groups with and without identified bacteria with respect to sex, race, clinical examination findings and laboratory results at admission, duration of antibiotic therapy before pleural fluid drainage, or length of stay (Table 1). The median pleural fluid white blood cell count was 6880 per mm3 (IQR, 154227,918 per mm3).

| Causative Bacteria Identified | |||
|---|---|---|---|
| Demographics | Yes (n = 11) | No (n = 53) | P Value |
| |||
| Female sex | 3 (27.3%) | 29 (54.7%) | 0.10 |
| Age (years) | 2 (1.34.50) | 4.3 (3.1 8.1) | 0.03 |
| Race | 0.09 | ||
| Non‐Hispanic white | 3 (27.3%) | 29 (54.7%) | |
| Non‐Hispanic black | 7 (63.6%) | 13 (24.5%) | |
| Hispanic | 0 (0.0%) | 3 (5.6%) | |
| Other | 1 (9.1%) | 8 (15.1%) | |
| History of asthma | 2 (18.2%%) | 12 (22.6%) | 0.75 |
| Duration of antibiotics before drainage (days) | 0.5 (03) | 2 (04) | 0.43 |
| Intensive care unit admission | 7 (63.6%) | 25 (47.2%) | 0.32 |
| Initial temperature (C) | 37.8 (37.239.5) | 38.1 (37.639.3) | 0.76 |
| Initial oxygen saturation (%) | 94 (9397) | 95 (9396) | 0.97 |
| Peripheral white blood cell count (per mm3) | 21.4 (14.028.4) | 16.3 (10.922.6) | 0.12 |
| Platelet count (per mm3) | 434 (298546) | 402 (312575) | 0.95 |
Blood was obtained for culture from 44 (68.8%) of 64 patients that underwent pleural fluid drainage. Blood cultures were positive in three (6.8%; 95% CI: 1.4%18.7%) of these patients; causative bacteria were S. pneumoniae (n = 1), Haemophilus influenzae (n = 1), and Staphylococcus aureus (n = 1). Pleural fluid cultures were positive from 6 of 64 patients (9.3%; 95% CI: 3.5%19.3%) that underwent pleural fluid drainage; causative bacteria were Staphylococcus aureus (n = 5) and Streptococcus pneumoniae (n = 1). Three of the 19 pleural fluid samples (15.8%, CI 3.4%39.6%) tested by 16S rRNA PCR yielded positive results; S. pneumoniae was identified by DNA sequencing in all three of these samples (Figure 1). Of these three specimens, two had both negative Gram stains and negative pleural fluid cultures, while one specimen had Gram‐positive cocci in pairs identified on Gram stain along with a pleural fluid culture‐positive for S. pneumoniae. Of the 16 patients with negative PCR results, one was found to have a pleural fluid culture‐positive for S. aureus; no bacteria were detected on the pleural fluid Gram stain (Figure 1). Differences in the demographic characteristics between patients in whom broad‐range PCR was performed and not performed were not statistically significant (Table 2). Differences in the median length of hospital stay between patients who had PCR performed (9 days; IQR, 712 days) and those who did not (11 days; IQR, 914 days) were not statistically significant (P = 0.19). Antibiotic therapy was simplified from treatment with multiple antibiotics to a single antibiotic in each instance of a positive culture, though the number of positive cultures was too small to lead to meaningful reductions in antibiotic use or spectrum.
| PCR Performed | |||
|---|---|---|---|
| Demographics | Yes (N = 19) | No (N = 45) | P Value |
| |||
| Female sex | 9 (47.4) | 23 (51.1) | 0.78 |
| Age (years) | 4.9 (2.78.8) | 4.0 (2.88.1) | 0.76 |
| Race | 0.42 | ||
| Non‐Hispanic white | 10 (52.6%) | 22 (48.9%) | |
| Non‐Hispanic black | 5 (26.3%) | 15 (33.3%) | |
| Hispanic | 0 (0.0%) | 3 (6.7%) | |
| Other | 4 (21.1%) | 5 (11.1%) | |
| History of asthma | 5 (26.32%) | 9 (20.0%) | 0.58 |
| Duration of antibiotics before drainage (days) | 2 (03) | 1.5 (04) | 0.65 |
| Intensive care unit admission | 8 (42.1%) | 24 (53.3%) | 0.41 |
| Initial temperature (C) | 38.1 (37.339.4) | 38.0 (37.639.4) | 0.86 |
| Initial pulse oximetry (%) | 96 (9497) | 94 (9296) | 0.34 |
| Initial white blood cell count (1000 per mm3) | 16.1 (9.527.4) | 17.8 (12.622.8) | 0.93 |
| Platelet count (1000 per mm3) | 397 (312575) | 431 (303597) | 0.73 |
Discussion
Identification of the causative organism can improve the treatment of children with complicated pneumonia by enabling clinicians to target the infection with effective, narrow‐spectrum antibiotics. However, broad‐spectrum antibiotics are typically given before blood or pleural fluid samples are obtained, lowering the yield of conventional bacterial cultures. In our study, pleural fluid and blood cultures were infrequently positive. However, the use of 16S rRNA broad‐range PCR and DNA sequencing as ancillary tests only modestly improved our diagnostic yield.
Two prior studies have shown that broad‐range PCR analysis of pleural fluid can detect pathogenic organisms from pleural fluid even after the administration of antibiotics. Saglani et al.12 used bacterial culture and broad‐range PCR to analyze pleural fluid from 32 children with complicated pneumonia. Although the cohort had received a median of eight days of antibiotic therapy prior to fluid aspiration, a combination of broad‐range PCR and DNA sequencing identified organisms, predominantly S. pneumoniae, in 17 of 26 (65.4%) pleural fluid samples with negative culture. Five of the six culture‐positive samples also had positive PCR results, suggesting that a low proportion of the PCR results were false‐negatives. Le Monnier et al.13 cultured the pleural fluid from 78 children with complicated pneumonia; 15 samples were excluded from PCR testing because they grew bacteria other than S. pneumoniae. Broad‐range 16S rRNA PCR detected bacteria in 22 of 40 (55.0%) samples that were culture‐negative; subsequent DNA sequencing identified S. pneumoniae (n = 17), S. pyogenes (n = 3), S. aureus (n = 1), and H. influenzae (n = 1). S. pneumoniae was identified by 16S rRNA PCR followed by DNA sequencing in 20 of the 23 (87.0%) pleural fluid samples that grew S. pneumoniae on culture.
PCR with organism‐specific primers has also been shown to detect pathogenic bacteria in 35%70% of pleural fluid samples from patients with complicated pneumonia.15, 16 These rates of detection are comparable to the results of prior studies that used broad‐range PCR, and suggest that 16S rRNA and organism‐specific PCR are similarly sensitive tests.
Our study found that the proportion of positive results with broad‐range PCR was greater than the proportion observed with conventional pleural fluid culture. We were also able to identify S. pneumoniae by broad‐range PCR in two culture‐negative pleural fluid samples. The detection of S. pneumoniae by PCR in the setting of negative pleural fluid culture is not surprising, as most patients received empiric antibiotic therapy against S. pneumoniae before undergoing pleural fluid drainage. This prior antibiotic therapy would be expected to decrease the yield of pleural fluid culture but have a less significant impact on PCR. The reason for the failure to detect S. aureus by PCR from our clinical samples is not known. We explored whether this issue could be attributable to test characteristics during validation of our assay (as described in the technical Appendix), and S. aureus was consistently identified in pleural fluid samples spiked with S. aureus. While our diagnostic yield was low compared with prior studies that used broad‐range PCR, the distribution of causative bacteria was similar to prior studies of complicated pneumonia.9, 10 There were too few positive cultures among the specimens available for broad‐range PCR testing for us to reliably assess 16S rRNA PCR sensitivity and specificity. Thus, it was not possible to reliably assess the value of 16S rRNA PCR as an ancillary test in culture‐negative complicated pneumonia.
Several factors may have contributed to the low yield of broad‐range PCR in our study. First, all patients in our study received broad‐spectrum antibiotics prior to pleural fluid drainage; most patients received treatment with a beta‐lactam agent in combination with either clindamycin or vancomycin. Although PCR is less affected than culture by the prior administration of antibiotics, it is still possible that exposure to antibiotics accelerated degradation of the bacterial genome, thus decreasing the sensitivity of broad‐range PCR. The median duration of antibiotic therapy prior to drainage was shorter for patients in whom a bacterial pathogen was identified, compared with those in whom a pathogen was not identified. Though this difference was not statistically significant, this difference emphasizes that early pleural fluid collection may improve bacterial detection. The median duration of prior antibiotic therapy was eight days, in the study by Saglani et al.,12 however details regarding anti‐staphylococcal therapy were not reported. Second, several studies using 16S rRNA PCR in blood or cerebrospinal fluid (CSF) specimens for children with sepsis or meningitis, respectively, have noted decreased sensitivity for detection of Gram‐positive bacteria.1720 The cell wall composition makes Gram‐positive bacteria particularly difficult to lyse for DNA extraction. Prior studies used a different approach to DNA extraction compared with our study. Saglani et al.12 used the QIAmp minikit for DNA extraction (Qiagen Ltd, West Sussex, UK) and Le Monnier et al.13 used the MagnaPure System (Roche Diagnostics, Indianapolis, IN) for DNA extraction; additionally Saglani et al.12 included an additional 15 minutes of incubation at 95C following Proteinase K digestion to ensure complete lysis of the bacterial cells. We used the MagMAX Total Nucleic Acid Isolation Kit with a bead beating step to degrade the samples prior to lysis. It is unlikely that any of these differences contributed to differences in our study results. Third, human DNA may cause a nonspecific background signal, which could decrease PCR sensitivity. This issue may be particularly important in complicated pneumonia where the pleural fluid white blood cell counts are substantially elevated. Human DNA from white blood cells might be present at much higher concentrations than bacterial DNA, which would create a competitive advantage for binding of human, rather than bacterial, DNA on to the bead matrix.21 Saglani et al.12 did not report pleural fluid white blood cell counts for comparison. However, Le Monnier et al.13 reported pleural fluid white blood cell counts slightly higher than those observed in our patients. Thus, differences in pleural fluid white blood cell counts do not necessarily explain the differences in yield across studies. Fourth, while the universal primers used in our study have been validated, including for detection of S. aureus, in previous studies,14 Saglani et al.12 and Le Monnier et al.13 each used a different set of primers. It is unclear whether or not the choice of different primers affected the microbiologic yield.
An important limitation of this study is the relatively small number of patients with pleural fluid available for PCR testing combined with the low PCR yield. This limitation may have caused us to underestimate the true benefit of 16S rRNA PCR as an ancillary diagnostic test, as suggested by the wide confidence intervals around the estimates of PCR yield. Additionally, not all patients underwent pleural drainage, and not all patients undergoing pleural drainage had pleural fluid available for PCR testing. The latter issue would likely have only minimal impact on our study results, as there were no differences in demographics or clinical or laboratory features among those with and without pleural fluid available for PCR testing. However, differences among physicians, and across institutions, may play an important role in the decision to perform pleural drainage. We do not know whether patients undergoing pleural drainage at our institution were more or less likely to have bacteria detected by PCR than patients not undergoing drainage. If they were more likely to have bacteria detected, then our study would underestimate the benefit of PCR as an adjunct diagnostic test. However, there was no difference in the duration of antibiotic pretreatment between these two groups, and the yield from blood culture was similar.
In conclusion, blood and pleural fluid cultures infrequently identify the causative bacteria in children with complicated pneumonia. The use of broad‐range PCR increased the microbiologic yield only modestly. Further refinements to improve the diagnostic accuracy of broad‐range PCR testing are needed before this technique can be recommended for widespread use in clinical practice.
Appendix
Optimization and Validation of The 16S Assay
The assay was optimized for use with pleural fluid samples as follows. E. coli (ATCC 25922) and S. aureus (ATCC 29213) were grown overnight in 100 ml Brain Heart Infusion (BHI) broth. A suspension equivalent to a 0.5 MacFarland standard was prepared and was taken to represent approximately 1 108 colony‐forming units/mL. To prepare standard PCR curves, 1 in 10 dilutions were prepared in PBS (pH, 7.4) and DNA was extracted, in triplicate, using two different DNA extraction methods: the MagMAX Total Nucleic Acid Isolation Kit and the PrepMan Ultra Sample Preparation reagent system (Applied Biosystems, Foster City, CA), as described by the manufacturer. For the MagMax extraction, a 300 microliter sample was added to 200 microliters of lysis/binding solution in a bead tube. Samples were disrupted using a vortex adapter for 15 minutes at the highest setting. Tubes were then centrifuged at 16,000 relative centrifugal force (RCF) for three minutes. DNA extraction was performed using the KingFisher Flex Automated Purification System (Thermo Scientific). For the PrepMan Ultra extraction, a 300‐l sample was centrifuged for three minutes at 16,000 RCF. The supernatant fluid was discarded and the pellet was resuspended in 100 microliters of PrepMan Ultra Lysis Buffer and incubated at 100C for ten minutes. The sample was centrifuged for three minutes at 16,000 RCF, and a 10‐microliter aliquot of supernatant fluid was added to 90 microliters of nuclease free water. All DNA extracts were frozen at 20C prior to use. Plate counts, performed from the bacterial suspensions in triplicate, revealed approximately 1.3 108 colony‐forming units/mL in the original suspensions.
Pleural fluid samples were spiked with known concentrations (1, 3, 10, 30, 100, 300, and 1000 colony‐forming units/mL) of E. coli and S. aureus, and each sample was extracted five times with both DNA extraction systems. Each DNA extract was amplified on an Applied Biosystems 7500 thermal cycler (ABI) and a SmartCycler (Cepheid, Sunnyvale, CA). Analysis of the results obtained from the preliminary experiments indicated that the following combination of DNA extraction and PCR was optimal, and these conditions were used for all patient samples.
DNA was extracted from a 300‐microliter aliquot of pleural fluid using the MagMAX Total Nucleic Acid Isolation Kit (Ambion, Applied Biosystems) as described by the manufacturer. PCR was performed on an Applied Biosystems 7500 thermal cycler (ABI) using the primers, 16S F: 5‐GCT GTC ATC TCT TGT GGG CTG T‐3, 16S R: 5‐AAA CTC ATG GGA GCT GCT GGT T‐3, and the fluorescent labeled TaqMan probe, 16s Probe: 5‐Cy5/CCT GTC ATG CCC ACA CAA ATC TCT CC/BHQ‐2‐3. Human DNA (0.5 nanograms) was added to each assay to ensure that an adequate amount of human DNA was available to act as an internal amplification control. The primers, AlbICF: 5‐GCT GTC ATC TCT TGT GGG CTG T‐3, AlbICR: 5‐AAA CTC ATG GGA GCT GCT GGT T‐3, and the TaqMan probe, AlbICP: 5‐Cy5/CCT GTC ATG CCC ACA CAA ATC TCT CC/BHQ‐2‐3 detect a region of the human albumin gene. Amplification was performed as follows: initial denaturation of 94C for 20 seconds, followed by 35 cycles of 94C for 10 seconds, 61C for 31 seconds, and 72C for 5 seconds. Each run included positive (E. coli ATCC 25922 DNA) and negative (nuclease free water) controls. A cycle threshold value of 30 or less indicated a positive sample. All positive samples were confirmed using the MicroSeq 500 16S rDNA Bacterial Identification kit (Applied Biosystems).
Community‐acquired pneumonia (CAP), the most common serious bacterial infection in childhood, may be complicated by parapneumonic effusion (ie, complicated pneumonia).1, 2 The clinical outcome of children with complicated pneumonia is directly influenced by the causative organism, and precise identification of the infectious agent has the potential to improve clinical management.35 The incidence of complicated pneumonia has recently increased, highlighting the need to better understand the reasons for this changing epidemiology.68 However, using conventional bacterial culture, bacteria are isolated from clinical samples obtained from children with complicated pneumonia in only 17%42% of cases.1, 9, 10 The low rate of positive culture results may be related to stringent bacterial growth requirements or to administration of broad‐spectrum antibiotics before obtaining blood or pleural fluid samples for culture.
Unlike bacterial culture, polymerase chain reaction (PCR) relies on the detection of bacterial DNA rather than on the recovery of viable bacteria and is therefore less affected by the prior administration of antibiotics. Broad‐range or universal primer 16S rRNA PCR detects a conserved region of the 16S ribosomal RNA (rRNA) gene and can detect a wide range of bacterial species with a single assay.11 However, few studies have evaluated the role of 16S rRNA PCR in detecting bacteria in the pleural fluid of children with complicated pneumonia.12, 13 The purpose of this study was to determine the frequency of positive blood and pleural fluid cultures in children with complicated pneumonia, and to determine whether broad‐range 16S rRNA PCR can increase the proportion of children with an identifiable microbiologic cause of complicated pneumonia.
Methods
Study Design, Setting, and Participants
This prospective cohort study was conducted at The Children's Hospital of Philadelphia (CHOP), an urban tertiary care children's hospital. The proposal was approved by the CHOP Committees for the Protection of Human Subjects. Patients were eligible for participation if they were 18 years of age, admitted to the hospital between October 1, 2007 and March 31, 2010, and diagnosed with complicated pneumonia. Parental informed consent was obtained for all patients, and verbal assent was obtained for all children over seven years of age. Patients with chronic medical conditions predisposing them to severe or recurrent pneumonia, such as human immunodeficiency virus, malignancy, and sickle cell disease, were excluded. The study team had no role in the clinical management of study patients. As this test is still considered experimental, pleural fluid 16S rRNA PCR was not performed until the patient was discharged from the hospital; these test results were not shared with the treating physicians.
Study Definition
Complicated pneumonia was defined by a temperature >38.0C, and the presence of lung parenchymal infiltrates and pleural effusions of any size or character on chest radiography or computed tomography.
Microbiologic MethodsConventional Culture
Pleural fluid or blood (4 mL) was inoculated onto a single pediatric BacT/Alert FAN bottle (BioMrieux, Durham, NC) which was immediately transported to the laboratory at room temperature. Additional pleural fluid was also submitted to the laboratory for a cytospin Gram stain. Once received in the laboratory, the bottles were immediately loaded into the BacT/Alert instrument. Bottles were automatically checked by the instrument for production of CO2 every ten minutes, and remained in the instrument for a total of five days. Bottles flagged as positive by the BacT/Alert system were removed from the instrument, subcultured to agar plates, and Gram stained. Bacterial isolates were identified and the antibiotic susceptibility tested by conventional methods following Clinical and Laboratory Standards Institute guidelines.
Microbiological Methods16S rRNA PCR
The 16S rRNA primers and probe used in this study have been validated previously and have been used extensively for the identification of organisms from the Domain Bacteria.14 Escherichia coli (ATCC 25922) and Staphylococcus aureus (ATCC 29213) were used as Gram‐negative and Gram‐positive controls, respectively. DNA extraction was performed on a 300 l aliquot of pleural fluid using the MagMAX Total Nucleic Acid Isolation Kit (Applied Biosystems, Foster City, CA). Samples were disrupted using a Vortex adapter for 15 minutes at the highest setting. Tubes were then centrifuged at 16,000 relative centrifugal force (RCF) for three minutes, and DNA extraction was performed using the KingFisher Flex Automated Purification System (Thermo Scientific, Waltham, MA). All DNA extracts were frozen at 20C prior to use. Details of the 16S rRNA PCR assay optimization and validation appear in the Appendix.
DNA was amplified on an Applied Biosystems 7500 thermal cycler (ABI, Foster City, CA) using the primers, 16S F: 5‐GCT GTC ATC TCT TGT GGG CTG T‐3, 16S R: 5‐AAA CTC ATG GGA GCT GCT GGT T‐3, and the fluorescent labeled TaqMan probe, 16s Probe: 5‐Cy5/CCT GTC ATG CCC ACA CAA ATC TCT CC/BHQ‐2‐3. Human DNA (0.5 nanograms) was added to each assay as a standardized internal amplification control. The primers, AlbICF: 5‐GCT GTC ATC TCT TGT GGG CTG T‐3, AlbICR: 5‐AAA CTC ATG GGA GCT GCT GGT T‐3, and the TaqMan probe, AlbICP: 5‐Cy5/CCT GTC ATG CCC ACA CAA ATC TCT CC/BHQ‐2‐3 detect a region of the human albumin gene. Amplification was performed as follows: initial denaturation of 94C for 20 seconds, followed by 35 cycles of 94C for 10 seconds, 61C for 31 seconds, and 72C for 5 seconds. Each run included positive (E. coli ATCC 25922 DNA) and negative (nuclease free water) controls. A cycle threshold value of 30 or less indicated a positive sample. All positive samples were confirmed using the MicroSeq 500 16S rDNA Bacterial Identification kit (Applied Biosystems).
DNA Sequencing
DNA sequencing was performed directly on PCR products using an ABI 3730 with the BigDye Taq FS Terminator V3.1. Sequencing was completed at the University of Pennsylvania DNA Sequencing Facility, and sequence identification was performed by BLAST at the National Center for Biotechnology Information web site (
Statistical Analysis
Data were analyzed using STATA 10.0 (Stata Corp., College Station, TX). Categorical variables were described using frequencies and percentages, and were compared using the chi‐square test. Continuous variables were described using mean, median, intraquartile range (IQR), and range values. Binomial exact 95% confidence intervals (CIs) were calculated for the proportion of positive blood and pleural fluid cultures and pleural fluid 16S rRNA PCR tests.
Results
During the study period, 124 patients with complicated pneumonia were identified; the median age was 4.9 years (IQR, 2.78.8 years) and the ratio of female to male patients was 1:1. The racial distribution was as follows: non‐Hispanic white, 62 (50%); non‐Hispanic black, 43 (34%); and Hispanic, 7 (6%). The median length of stay was 11 days (range, 240 days; IQR, 811 days). Forty‐one (33.1%) patients were admitted to the intensive care unit, and nine (7.3%) patients required mechanical ventilation. Overall, 5 of the 71 blood cultures performed (7.0%; 95% CI: 2.3%15.7%) were positive.
Pleural fluid drainage was performed in 64 (51.6%) patients. The median duration of antibiotic treatment before pleural fluid drainage was two days (IQR, 04 days). Differences in duration of antibiotic pretreatment between those with and without pleural fluid drainage were not significant. Patients received beta‐lactam agents (76.6%), clindamycin (51.6%), and vancomycin (23.4%) before pleural drainage. Blood cultures were performed in 27 (45.0%) of 60 patients not undergoing pleural fluid drainage; 2 of these 27 (7.4%; 95% CI: 0.9%24.3%) blood cultures were positive, and Streptococcus pneumoniae was isolated in both cases.
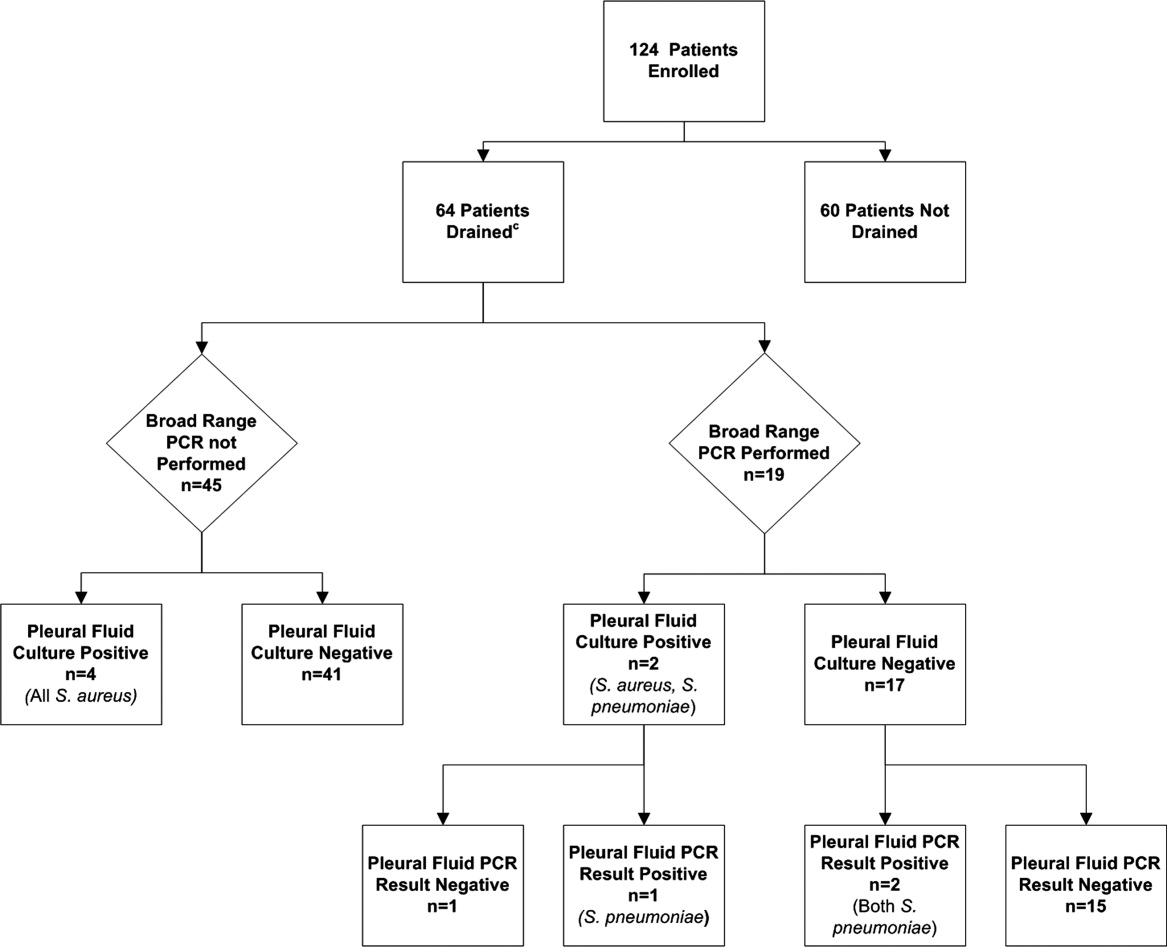
Using a combination of pleural fluid culture, blood culture, and broad‐range PCR, a microbiologic cause of infection was identified in 11 of 64 (17.2%; 95% CI: 8.9%28.7%) patients undergoing pleural fluid drainage (Figure 1). Patients with an identified bacterial cause were younger than those without an identified bacterial cause (Table 1). However, there were no statistically significant differences between the groups with and without identified bacteria with respect to sex, race, clinical examination findings and laboratory results at admission, duration of antibiotic therapy before pleural fluid drainage, or length of stay (Table 1). The median pleural fluid white blood cell count was 6880 per mm3 (IQR, 154227,918 per mm3).

| Causative Bacteria Identified | |||
|---|---|---|---|
| Demographics | Yes (n = 11) | No (n = 53) | P Value |
| |||
| Female sex | 3 (27.3%) | 29 (54.7%) | 0.10 |
| Age (years) | 2 (1.34.50) | 4.3 (3.1 8.1) | 0.03 |
| Race | 0.09 | ||
| Non‐Hispanic white | 3 (27.3%) | 29 (54.7%) | |
| Non‐Hispanic black | 7 (63.6%) | 13 (24.5%) | |
| Hispanic | 0 (0.0%) | 3 (5.6%) | |
| Other | 1 (9.1%) | 8 (15.1%) | |
| History of asthma | 2 (18.2%%) | 12 (22.6%) | 0.75 |
| Duration of antibiotics before drainage (days) | 0.5 (03) | 2 (04) | 0.43 |
| Intensive care unit admission | 7 (63.6%) | 25 (47.2%) | 0.32 |
| Initial temperature (C) | 37.8 (37.239.5) | 38.1 (37.639.3) | 0.76 |
| Initial oxygen saturation (%) | 94 (9397) | 95 (9396) | 0.97 |
| Peripheral white blood cell count (per mm3) | 21.4 (14.028.4) | 16.3 (10.922.6) | 0.12 |
| Platelet count (per mm3) | 434 (298546) | 402 (312575) | 0.95 |
Blood was obtained for culture from 44 (68.8%) of 64 patients that underwent pleural fluid drainage. Blood cultures were positive in three (6.8%; 95% CI: 1.4%18.7%) of these patients; causative bacteria were S. pneumoniae (n = 1), Haemophilus influenzae (n = 1), and Staphylococcus aureus (n = 1). Pleural fluid cultures were positive from 6 of 64 patients (9.3%; 95% CI: 3.5%19.3%) that underwent pleural fluid drainage; causative bacteria were Staphylococcus aureus (n = 5) and Streptococcus pneumoniae (n = 1). Three of the 19 pleural fluid samples (15.8%, CI 3.4%39.6%) tested by 16S rRNA PCR yielded positive results; S. pneumoniae was identified by DNA sequencing in all three of these samples (Figure 1). Of these three specimens, two had both negative Gram stains and negative pleural fluid cultures, while one specimen had Gram‐positive cocci in pairs identified on Gram stain along with a pleural fluid culture‐positive for S. pneumoniae. Of the 16 patients with negative PCR results, one was found to have a pleural fluid culture‐positive for S. aureus; no bacteria were detected on the pleural fluid Gram stain (Figure 1). Differences in the demographic characteristics between patients in whom broad‐range PCR was performed and not performed were not statistically significant (Table 2). Differences in the median length of hospital stay between patients who had PCR performed (9 days; IQR, 712 days) and those who did not (11 days; IQR, 914 days) were not statistically significant (P = 0.19). Antibiotic therapy was simplified from treatment with multiple antibiotics to a single antibiotic in each instance of a positive culture, though the number of positive cultures was too small to lead to meaningful reductions in antibiotic use or spectrum.
| PCR Performed | |||
|---|---|---|---|
| Demographics | Yes (N = 19) | No (N = 45) | P Value |
| |||
| Female sex | 9 (47.4) | 23 (51.1) | 0.78 |
| Age (years) | 4.9 (2.78.8) | 4.0 (2.88.1) | 0.76 |
| Race | 0.42 | ||
| Non‐Hispanic white | 10 (52.6%) | 22 (48.9%) | |
| Non‐Hispanic black | 5 (26.3%) | 15 (33.3%) | |
| Hispanic | 0 (0.0%) | 3 (6.7%) | |
| Other | 4 (21.1%) | 5 (11.1%) | |
| History of asthma | 5 (26.32%) | 9 (20.0%) | 0.58 |
| Duration of antibiotics before drainage (days) | 2 (03) | 1.5 (04) | 0.65 |
| Intensive care unit admission | 8 (42.1%) | 24 (53.3%) | 0.41 |
| Initial temperature (C) | 38.1 (37.339.4) | 38.0 (37.639.4) | 0.86 |
| Initial pulse oximetry (%) | 96 (9497) | 94 (9296) | 0.34 |
| Initial white blood cell count (1000 per mm3) | 16.1 (9.527.4) | 17.8 (12.622.8) | 0.93 |
| Platelet count (1000 per mm3) | 397 (312575) | 431 (303597) | 0.73 |
Discussion
Identification of the causative organism can improve the treatment of children with complicated pneumonia by enabling clinicians to target the infection with effective, narrow‐spectrum antibiotics. However, broad‐spectrum antibiotics are typically given before blood or pleural fluid samples are obtained, lowering the yield of conventional bacterial cultures. In our study, pleural fluid and blood cultures were infrequently positive. However, the use of 16S rRNA broad‐range PCR and DNA sequencing as ancillary tests only modestly improved our diagnostic yield.
Two prior studies have shown that broad‐range PCR analysis of pleural fluid can detect pathogenic organisms from pleural fluid even after the administration of antibiotics. Saglani et al.12 used bacterial culture and broad‐range PCR to analyze pleural fluid from 32 children with complicated pneumonia. Although the cohort had received a median of eight days of antibiotic therapy prior to fluid aspiration, a combination of broad‐range PCR and DNA sequencing identified organisms, predominantly S. pneumoniae, in 17 of 26 (65.4%) pleural fluid samples with negative culture. Five of the six culture‐positive samples also had positive PCR results, suggesting that a low proportion of the PCR results were false‐negatives. Le Monnier et al.13 cultured the pleural fluid from 78 children with complicated pneumonia; 15 samples were excluded from PCR testing because they grew bacteria other than S. pneumoniae. Broad‐range 16S rRNA PCR detected bacteria in 22 of 40 (55.0%) samples that were culture‐negative; subsequent DNA sequencing identified S. pneumoniae (n = 17), S. pyogenes (n = 3), S. aureus (n = 1), and H. influenzae (n = 1). S. pneumoniae was identified by 16S rRNA PCR followed by DNA sequencing in 20 of the 23 (87.0%) pleural fluid samples that grew S. pneumoniae on culture.
PCR with organism‐specific primers has also been shown to detect pathogenic bacteria in 35%70% of pleural fluid samples from patients with complicated pneumonia.15, 16 These rates of detection are comparable to the results of prior studies that used broad‐range PCR, and suggest that 16S rRNA and organism‐specific PCR are similarly sensitive tests.
Our study found that the proportion of positive results with broad‐range PCR was greater than the proportion observed with conventional pleural fluid culture. We were also able to identify S. pneumoniae by broad‐range PCR in two culture‐negative pleural fluid samples. The detection of S. pneumoniae by PCR in the setting of negative pleural fluid culture is not surprising, as most patients received empiric antibiotic therapy against S. pneumoniae before undergoing pleural fluid drainage. This prior antibiotic therapy would be expected to decrease the yield of pleural fluid culture but have a less significant impact on PCR. The reason for the failure to detect S. aureus by PCR from our clinical samples is not known. We explored whether this issue could be attributable to test characteristics during validation of our assay (as described in the technical Appendix), and S. aureus was consistently identified in pleural fluid samples spiked with S. aureus. While our diagnostic yield was low compared with prior studies that used broad‐range PCR, the distribution of causative bacteria was similar to prior studies of complicated pneumonia.9, 10 There were too few positive cultures among the specimens available for broad‐range PCR testing for us to reliably assess 16S rRNA PCR sensitivity and specificity. Thus, it was not possible to reliably assess the value of 16S rRNA PCR as an ancillary test in culture‐negative complicated pneumonia.
Several factors may have contributed to the low yield of broad‐range PCR in our study. First, all patients in our study received broad‐spectrum antibiotics prior to pleural fluid drainage; most patients received treatment with a beta‐lactam agent in combination with either clindamycin or vancomycin. Although PCR is less affected than culture by the prior administration of antibiotics, it is still possible that exposure to antibiotics accelerated degradation of the bacterial genome, thus decreasing the sensitivity of broad‐range PCR. The median duration of antibiotic therapy prior to drainage was shorter for patients in whom a bacterial pathogen was identified, compared with those in whom a pathogen was not identified. Though this difference was not statistically significant, this difference emphasizes that early pleural fluid collection may improve bacterial detection. The median duration of prior antibiotic therapy was eight days, in the study by Saglani et al.,12 however details regarding anti‐staphylococcal therapy were not reported. Second, several studies using 16S rRNA PCR in blood or cerebrospinal fluid (CSF) specimens for children with sepsis or meningitis, respectively, have noted decreased sensitivity for detection of Gram‐positive bacteria.1720 The cell wall composition makes Gram‐positive bacteria particularly difficult to lyse for DNA extraction. Prior studies used a different approach to DNA extraction compared with our study. Saglani et al.12 used the QIAmp minikit for DNA extraction (Qiagen Ltd, West Sussex, UK) and Le Monnier et al.13 used the MagnaPure System (Roche Diagnostics, Indianapolis, IN) for DNA extraction; additionally Saglani et al.12 included an additional 15 minutes of incubation at 95C following Proteinase K digestion to ensure complete lysis of the bacterial cells. We used the MagMAX Total Nucleic Acid Isolation Kit with a bead beating step to degrade the samples prior to lysis. It is unlikely that any of these differences contributed to differences in our study results. Third, human DNA may cause a nonspecific background signal, which could decrease PCR sensitivity. This issue may be particularly important in complicated pneumonia where the pleural fluid white blood cell counts are substantially elevated. Human DNA from white blood cells might be present at much higher concentrations than bacterial DNA, which would create a competitive advantage for binding of human, rather than bacterial, DNA on to the bead matrix.21 Saglani et al.12 did not report pleural fluid white blood cell counts for comparison. However, Le Monnier et al.13 reported pleural fluid white blood cell counts slightly higher than those observed in our patients. Thus, differences in pleural fluid white blood cell counts do not necessarily explain the differences in yield across studies. Fourth, while the universal primers used in our study have been validated, including for detection of S. aureus, in previous studies,14 Saglani et al.12 and Le Monnier et al.13 each used a different set of primers. It is unclear whether or not the choice of different primers affected the microbiologic yield.
An important limitation of this study is the relatively small number of patients with pleural fluid available for PCR testing combined with the low PCR yield. This limitation may have caused us to underestimate the true benefit of 16S rRNA PCR as an ancillary diagnostic test, as suggested by the wide confidence intervals around the estimates of PCR yield. Additionally, not all patients underwent pleural drainage, and not all patients undergoing pleural drainage had pleural fluid available for PCR testing. The latter issue would likely have only minimal impact on our study results, as there were no differences in demographics or clinical or laboratory features among those with and without pleural fluid available for PCR testing. However, differences among physicians, and across institutions, may play an important role in the decision to perform pleural drainage. We do not know whether patients undergoing pleural drainage at our institution were more or less likely to have bacteria detected by PCR than patients not undergoing drainage. If they were more likely to have bacteria detected, then our study would underestimate the benefit of PCR as an adjunct diagnostic test. However, there was no difference in the duration of antibiotic pretreatment between these two groups, and the yield from blood culture was similar.
In conclusion, blood and pleural fluid cultures infrequently identify the causative bacteria in children with complicated pneumonia. The use of broad‐range PCR increased the microbiologic yield only modestly. Further refinements to improve the diagnostic accuracy of broad‐range PCR testing are needed before this technique can be recommended for widespread use in clinical practice.
Appendix
Optimization and Validation of The 16S Assay
The assay was optimized for use with pleural fluid samples as follows. E. coli (ATCC 25922) and S. aureus (ATCC 29213) were grown overnight in 100 ml Brain Heart Infusion (BHI) broth. A suspension equivalent to a 0.5 MacFarland standard was prepared and was taken to represent approximately 1 108 colony‐forming units/mL. To prepare standard PCR curves, 1 in 10 dilutions were prepared in PBS (pH, 7.4) and DNA was extracted, in triplicate, using two different DNA extraction methods: the MagMAX Total Nucleic Acid Isolation Kit and the PrepMan Ultra Sample Preparation reagent system (Applied Biosystems, Foster City, CA), as described by the manufacturer. For the MagMax extraction, a 300 microliter sample was added to 200 microliters of lysis/binding solution in a bead tube. Samples were disrupted using a vortex adapter for 15 minutes at the highest setting. Tubes were then centrifuged at 16,000 relative centrifugal force (RCF) for three minutes. DNA extraction was performed using the KingFisher Flex Automated Purification System (Thermo Scientific). For the PrepMan Ultra extraction, a 300‐l sample was centrifuged for three minutes at 16,000 RCF. The supernatant fluid was discarded and the pellet was resuspended in 100 microliters of PrepMan Ultra Lysis Buffer and incubated at 100C for ten minutes. The sample was centrifuged for three minutes at 16,000 RCF, and a 10‐microliter aliquot of supernatant fluid was added to 90 microliters of nuclease free water. All DNA extracts were frozen at 20C prior to use. Plate counts, performed from the bacterial suspensions in triplicate, revealed approximately 1.3 108 colony‐forming units/mL in the original suspensions.
Pleural fluid samples were spiked with known concentrations (1, 3, 10, 30, 100, 300, and 1000 colony‐forming units/mL) of E. coli and S. aureus, and each sample was extracted five times with both DNA extraction systems. Each DNA extract was amplified on an Applied Biosystems 7500 thermal cycler (ABI) and a SmartCycler (Cepheid, Sunnyvale, CA). Analysis of the results obtained from the preliminary experiments indicated that the following combination of DNA extraction and PCR was optimal, and these conditions were used for all patient samples.
DNA was extracted from a 300‐microliter aliquot of pleural fluid using the MagMAX Total Nucleic Acid Isolation Kit (Ambion, Applied Biosystems) as described by the manufacturer. PCR was performed on an Applied Biosystems 7500 thermal cycler (ABI) using the primers, 16S F: 5‐GCT GTC ATC TCT TGT GGG CTG T‐3, 16S R: 5‐AAA CTC ATG GGA GCT GCT GGT T‐3, and the fluorescent labeled TaqMan probe, 16s Probe: 5‐Cy5/CCT GTC ATG CCC ACA CAA ATC TCT CC/BHQ‐2‐3. Human DNA (0.5 nanograms) was added to each assay to ensure that an adequate amount of human DNA was available to act as an internal amplification control. The primers, AlbICF: 5‐GCT GTC ATC TCT TGT GGG CTG T‐3, AlbICR: 5‐AAA CTC ATG GGA GCT GCT GGT T‐3, and the TaqMan probe, AlbICP: 5‐Cy5/CCT GTC ATG CCC ACA CAA ATC TCT CC/BHQ‐2‐3 detect a region of the human albumin gene. Amplification was performed as follows: initial denaturation of 94C for 20 seconds, followed by 35 cycles of 94C for 10 seconds, 61C for 31 seconds, and 72C for 5 seconds. Each run included positive (E. coli ATCC 25922 DNA) and negative (nuclease free water) controls. A cycle threshold value of 30 or less indicated a positive sample. All positive samples were confirmed using the MicroSeq 500 16S rDNA Bacterial Identification kit (Applied Biosystems).
- ,,, et al.An epidemiological investigation of a sustained high rate of pediatric parapneumonic empyema: risk factors and microbiological associations.Clin Infect Dis.2002;34:434–440.
- ,,,,.Primary early thoracoscopy and reduction in length of hospital stay and additional procedures among children with complicated pneumonia: results of a multicenter retrospective cohort study.Arch Pediatr Adolesc Med.2008;162:675–681.
- ,,, et al.Pulmonary manifestations in children with invasive community‐acquired Staphylococcus aureus infection.Clin Infect Dis.2005;41:583–590.
- ,,, et al.Association between Staphylococcus aureus strains carrying gene for Panton‐Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients.Lancet.2002;359:753–759.
- ,,, et al.Severe community‐onset pneumonia in healthy adults caused by methicillin‐resistant Staphylococcus aureus carrying the Panton‐Valentine leukocidin genes.Clin Infect Dis.2005;40:100–107.
- ,.Empyema hospitalizations increased in US children despite pneumococcal conjugate vaccine.Pediatrics.2010;125:26–33.
- ,,,.Increasing incidence of empyema complicating childhood community‐acquired pneumonia in the United States.Clin Infect Dis.2010;50:805–813.
- ,,,,.National hospitalization trends for pediatric pneumonia and associated complications.Pediatrics.2010;126:204–213.
- ,,, et al.Thoracoscopic decortication vs tube thoracostomy with fibrinolysis for empyema in children: a prospective, randomized trial.J Pediatr Surg.2009;44:106–111.
- ,,, et al.Comparison of urokinase and video‐assisted thoracoscopic surgery for treatment of childhood empyema.Am J Respir Crit Care Med.2006;174:221–227.
- ,.Development of broad‐range 16S rDNA PCR for use in the routine diagnostic clinical microbiology service.J Med Microbiol.2003;52:685–691.
- ,,,.Empyema: the use of broad range 16S rDNA PCR for pathogen detection.Arch Dis Child.2005;90:70–73.
- ,,, et al.Microbiological diagnosis of empyema in children: comparative evaluations by culture, polymerase chain reaction, and pneumococcal antigen detection in pleural fluids.Clin Infect Dis.2006;42:1135–1140.
- ,,,.Determination of bacterial load by real‐time PCR using a broad‐range (universal) probe and primers set.Microbiology.2002;148:257–266.
- ,,,,,.Pneumolysin polymerase chain reaction for diagnosis of pneumococcal pneumonia and empyema in children.Eur J Clin Microbiol Infect Dis.2006;25:783–789.
- ,,, et al.Pleural fluid PCR method for detection of Staphylococcus aureus, Streptococcus pneumoniae and Haemophilus influenzae in pediatric parapneumonic effusions.Respiration.2008;75:437–442.
- ,.16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls.J Clin Microbiol.2007;45:2761–2764.
- ,,,,.Evaluating the near‐term infant for early onset sepsis: progress and challenges to consider with 16S rDNA polymerase chain reaction testing.J Mol Diagn.2006;8:357–363.
- ,,,,.Preanalytic removal of human DNA eliminates false signals in general 16S rDNA PCR monitoring of bacterial pathogens in blood.Comp Immunol Microbiol Infect Dis.2009;32:207–219.
- ,,,,.Use of broad range16S rDNA PCR in clinical microbiology.J Microbiol Methods.2009;76:217–225.
- ,.Real‐time polymerase chain reaction for detecting bacterial DNA directly from blood of neonates being evaluated for sepsis.J Mol Diagn.2005;7:575–581.
- ,,, et al.An epidemiological investigation of a sustained high rate of pediatric parapneumonic empyema: risk factors and microbiological associations.Clin Infect Dis.2002;34:434–440.
- ,,,,.Primary early thoracoscopy and reduction in length of hospital stay and additional procedures among children with complicated pneumonia: results of a multicenter retrospective cohort study.Arch Pediatr Adolesc Med.2008;162:675–681.
- ,,, et al.Pulmonary manifestations in children with invasive community‐acquired Staphylococcus aureus infection.Clin Infect Dis.2005;41:583–590.
- ,,, et al.Association between Staphylococcus aureus strains carrying gene for Panton‐Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients.Lancet.2002;359:753–759.
- ,,, et al.Severe community‐onset pneumonia in healthy adults caused by methicillin‐resistant Staphylococcus aureus carrying the Panton‐Valentine leukocidin genes.Clin Infect Dis.2005;40:100–107.
- ,.Empyema hospitalizations increased in US children despite pneumococcal conjugate vaccine.Pediatrics.2010;125:26–33.
- ,,,.Increasing incidence of empyema complicating childhood community‐acquired pneumonia in the United States.Clin Infect Dis.2010;50:805–813.
- ,,,,.National hospitalization trends for pediatric pneumonia and associated complications.Pediatrics.2010;126:204–213.
- ,,, et al.Thoracoscopic decortication vs tube thoracostomy with fibrinolysis for empyema in children: a prospective, randomized trial.J Pediatr Surg.2009;44:106–111.
- ,,, et al.Comparison of urokinase and video‐assisted thoracoscopic surgery for treatment of childhood empyema.Am J Respir Crit Care Med.2006;174:221–227.
- ,.Development of broad‐range 16S rDNA PCR for use in the routine diagnostic clinical microbiology service.J Med Microbiol.2003;52:685–691.
- ,,,.Empyema: the use of broad range 16S rDNA PCR for pathogen detection.Arch Dis Child.2005;90:70–73.
- ,,, et al.Microbiological diagnosis of empyema in children: comparative evaluations by culture, polymerase chain reaction, and pneumococcal antigen detection in pleural fluids.Clin Infect Dis.2006;42:1135–1140.
- ,,,.Determination of bacterial load by real‐time PCR using a broad‐range (universal) probe and primers set.Microbiology.2002;148:257–266.
- ,,,,,.Pneumolysin polymerase chain reaction for diagnosis of pneumococcal pneumonia and empyema in children.Eur J Clin Microbiol Infect Dis.2006;25:783–789.
- ,,, et al.Pleural fluid PCR method for detection of Staphylococcus aureus, Streptococcus pneumoniae and Haemophilus influenzae in pediatric parapneumonic effusions.Respiration.2008;75:437–442.
- ,.16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls.J Clin Microbiol.2007;45:2761–2764.
- ,,,,.Evaluating the near‐term infant for early onset sepsis: progress and challenges to consider with 16S rDNA polymerase chain reaction testing.J Mol Diagn.2006;8:357–363.
- ,,,,.Preanalytic removal of human DNA eliminates false signals in general 16S rDNA PCR monitoring of bacterial pathogens in blood.Comp Immunol Microbiol Infect Dis.2009;32:207–219.
- ,,,,.Use of broad range16S rDNA PCR in clinical microbiology.J Microbiol Methods.2009;76:217–225.
- ,.Real‐time polymerase chain reaction for detecting bacterial DNA directly from blood of neonates being evaluated for sepsis.J Mol Diagn.2005;7:575–581.
Copyright © 2011 Society of Hospital Medicine