User login
Researchers have identified the structure of a protein that could turn out to be a potential vaccine target for the 2019-nCoV.
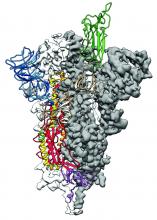
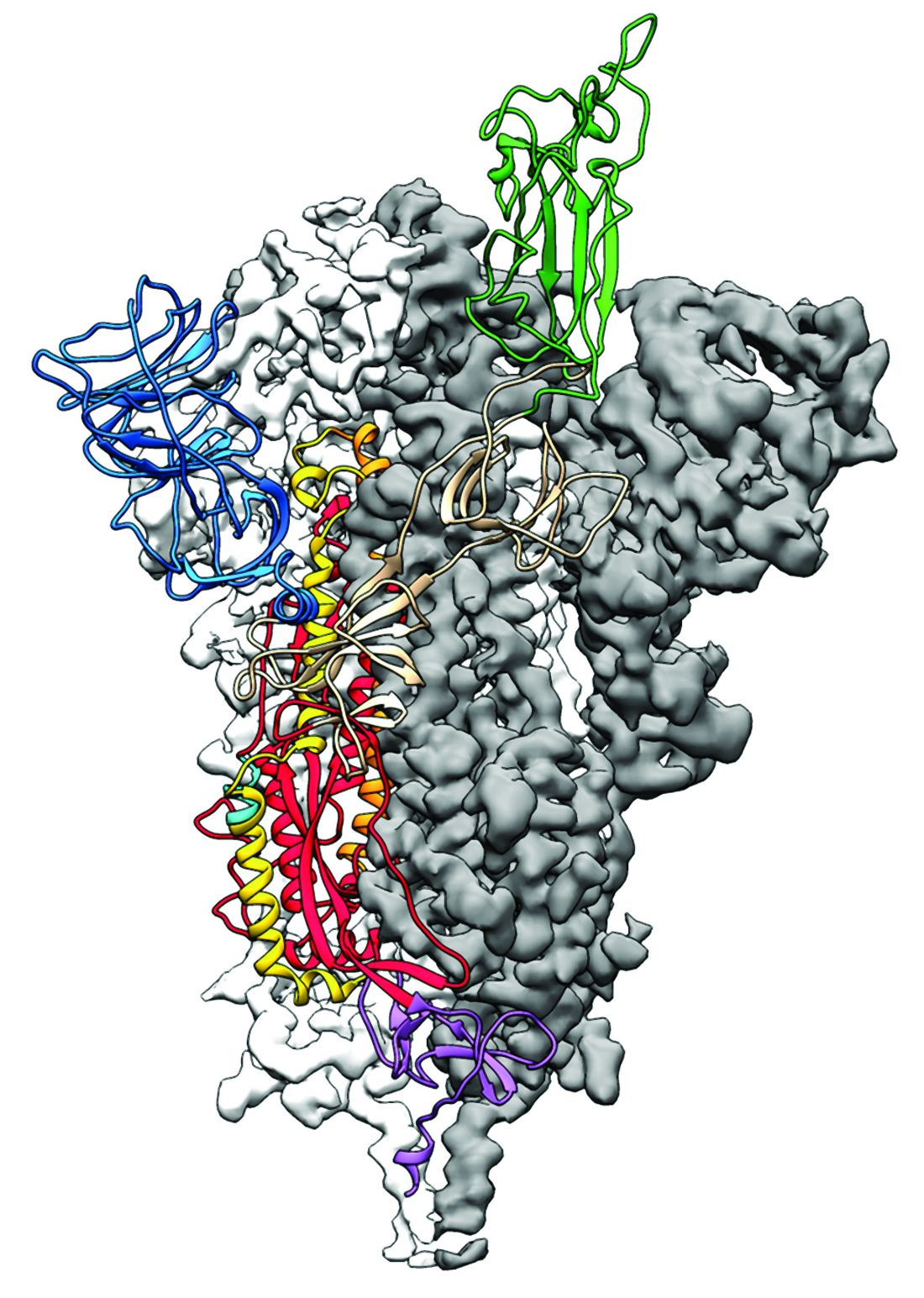
As is typical of other coronaviruses, 2019-nCoV makes use of a densely glycosylated spike protein to gain entry into host cells. The spike protein is a trimeric class I fusion protein that exists in a metastable prefusion conformation that undergoes a dramatic structural rearrangement to fuse the viral membrane with the host-cell membrane, according to Daniel Wrapp of the University of Texas at Austin and colleagues.
The researchers performed a study to synthesize and determine the 3-D structure of the spike protein because it is a logical target for vaccine development and for the development of targeted therapeutics for COVID-19, the disease caused by the virus.
“As soon as we knew this was a coronavirus, we felt we had to jump at it,” senior author Jason S. McLellan, PhD, associate professor of molecular science, said in a press release from the University, “because we could be one of the first ones to get this structure. We knew exactly what mutations to put into this because we’ve already shown these mutations work for a bunch of other coronaviruses.”
Because recent reports by other researchers demonstrated that 2019-nCoV and SARS-CoV spike proteins share the same functional host-cell receptor–angiotensin-converting enzyme 2 (ACE2), Dr. McLellan and his colleagues examined the relation between the two viruses. They found biophysical and structural evidence that the 2019-nCoV spike protein binds ACE2 with higher affinity than the closely related SARS-CoV spike protein. “The high affinity of 2019-nCoV S for human ACE2 may contribute to the apparent ease with which 2019-nCoV can spread from human-to-human; however, additional studies are needed to investigate this possibility,” the researchers wrote.
Focusing their attention on the receptor-binding domain (RBD) of the 2019-nCoV spike protein, they tested several published SARS-CoV RBD-specific monoclonal antibodies against it and found that these antibodies showed no appreciable binding to 2019-nCoV spike protein, which suggests limited antibody cross-reactivity. For this reason, they suggested that future antibody isolation and therapeutic design efforts will benefit from specifically using 2019-nCoV spike proteins as probes.
“This information will support precision vaccine design and discovery of anti-viral therapeutics, accelerating medical countermeasure development,” they concluded.
The research was supported in part by an National Institutes of Health/National Institute of Allergy and Infectious Diseases grant and by intramural funding from the National Institute of Allergy and Infectious Diseases. Four authors are inventors on US patent application No. 62/412,703 (Prefusion Coronavirus Spike Proteins and Their Use) and all are inventors on US patent application No. 62/972,886 (2019-nCoV Vaccine).
SOURCE: Wrapp D et al. Science. 2020 Feb 19. doi: 10.1126/science.abb2507.
Researchers have identified the structure of a protein that could turn out to be a potential vaccine target for the 2019-nCoV.
As is typical of other coronaviruses, 2019-nCoV makes use of a densely glycosylated spike protein to gain entry into host cells. The spike protein is a trimeric class I fusion protein that exists in a metastable prefusion conformation that undergoes a dramatic structural rearrangement to fuse the viral membrane with the host-cell membrane, according to Daniel Wrapp of the University of Texas at Austin and colleagues.
The researchers performed a study to synthesize and determine the 3-D structure of the spike protein because it is a logical target for vaccine development and for the development of targeted therapeutics for COVID-19, the disease caused by the virus.
“As soon as we knew this was a coronavirus, we felt we had to jump at it,” senior author Jason S. McLellan, PhD, associate professor of molecular science, said in a press release from the University, “because we could be one of the first ones to get this structure. We knew exactly what mutations to put into this because we’ve already shown these mutations work for a bunch of other coronaviruses.”
Because recent reports by other researchers demonstrated that 2019-nCoV and SARS-CoV spike proteins share the same functional host-cell receptor–angiotensin-converting enzyme 2 (ACE2), Dr. McLellan and his colleagues examined the relation between the two viruses. They found biophysical and structural evidence that the 2019-nCoV spike protein binds ACE2 with higher affinity than the closely related SARS-CoV spike protein. “The high affinity of 2019-nCoV S for human ACE2 may contribute to the apparent ease with which 2019-nCoV can spread from human-to-human; however, additional studies are needed to investigate this possibility,” the researchers wrote.
Focusing their attention on the receptor-binding domain (RBD) of the 2019-nCoV spike protein, they tested several published SARS-CoV RBD-specific monoclonal antibodies against it and found that these antibodies showed no appreciable binding to 2019-nCoV spike protein, which suggests limited antibody cross-reactivity. For this reason, they suggested that future antibody isolation and therapeutic design efforts will benefit from specifically using 2019-nCoV spike proteins as probes.
“This information will support precision vaccine design and discovery of anti-viral therapeutics, accelerating medical countermeasure development,” they concluded.
The research was supported in part by an National Institutes of Health/National Institute of Allergy and Infectious Diseases grant and by intramural funding from the National Institute of Allergy and Infectious Diseases. Four authors are inventors on US patent application No. 62/412,703 (Prefusion Coronavirus Spike Proteins and Their Use) and all are inventors on US patent application No. 62/972,886 (2019-nCoV Vaccine).
SOURCE: Wrapp D et al. Science. 2020 Feb 19. doi: 10.1126/science.abb2507.
Researchers have identified the structure of a protein that could turn out to be a potential vaccine target for the 2019-nCoV.
As is typical of other coronaviruses, 2019-nCoV makes use of a densely glycosylated spike protein to gain entry into host cells. The spike protein is a trimeric class I fusion protein that exists in a metastable prefusion conformation that undergoes a dramatic structural rearrangement to fuse the viral membrane with the host-cell membrane, according to Daniel Wrapp of the University of Texas at Austin and colleagues.
The researchers performed a study to synthesize and determine the 3-D structure of the spike protein because it is a logical target for vaccine development and for the development of targeted therapeutics for COVID-19, the disease caused by the virus.
“As soon as we knew this was a coronavirus, we felt we had to jump at it,” senior author Jason S. McLellan, PhD, associate professor of molecular science, said in a press release from the University, “because we could be one of the first ones to get this structure. We knew exactly what mutations to put into this because we’ve already shown these mutations work for a bunch of other coronaviruses.”
Because recent reports by other researchers demonstrated that 2019-nCoV and SARS-CoV spike proteins share the same functional host-cell receptor–angiotensin-converting enzyme 2 (ACE2), Dr. McLellan and his colleagues examined the relation between the two viruses. They found biophysical and structural evidence that the 2019-nCoV spike protein binds ACE2 with higher affinity than the closely related SARS-CoV spike protein. “The high affinity of 2019-nCoV S for human ACE2 may contribute to the apparent ease with which 2019-nCoV can spread from human-to-human; however, additional studies are needed to investigate this possibility,” the researchers wrote.
Focusing their attention on the receptor-binding domain (RBD) of the 2019-nCoV spike protein, they tested several published SARS-CoV RBD-specific monoclonal antibodies against it and found that these antibodies showed no appreciable binding to 2019-nCoV spike protein, which suggests limited antibody cross-reactivity. For this reason, they suggested that future antibody isolation and therapeutic design efforts will benefit from specifically using 2019-nCoV spike proteins as probes.
“This information will support precision vaccine design and discovery of anti-viral therapeutics, accelerating medical countermeasure development,” they concluded.
The research was supported in part by an National Institutes of Health/National Institute of Allergy and Infectious Diseases grant and by intramural funding from the National Institute of Allergy and Infectious Diseases. Four authors are inventors on US patent application No. 62/412,703 (Prefusion Coronavirus Spike Proteins and Their Use) and all are inventors on US patent application No. 62/972,886 (2019-nCoV Vaccine).
SOURCE: Wrapp D et al. Science. 2020 Feb 19. doi: 10.1126/science.abb2507.
FROM SCIENCE