User login
Pathologic and, increasingly, molecular analysis of tumor tissue biopsies is the gold standard in initial diagnosis of cancer, but liquid biopsies, which analyze tumor-derived material circulating in the bloodstream are gaining traction. Here, we discuss the current state of development of this complementary and potentially alternative approach to tumor analysis.
Liquid biopsy gaining traction
Biopsies enable oncologists to gather information about a potential or established tumor, including confirmation of the presence of cancerous tissue and determination of its histological characteristics, such as tumor grade and stage, as well as its molecular features, such as the presence of certain gene mutations. Ultimately, this information can be put to use in determining the most appropriate course of treatment.
The current gold standard is a tissue biopsy that typically involves an invasive procedure to permit the collection of a piece of tumor tissue. Yet, tissue biopsies are not always feasible because of the location of the tumor or the poor performance status of many patients with advanced disease. They also provide only a snapshot of the disease at the time at which they were taken and don’t necessarily reflect the genetic heterogeneity or evolution of a tumor over time.
The detection of components that are derived from the tumor circulating in the blood of cancer patients had fueled the idea of blood-based diagnostics in oncology – so-called liquid biopsies. These have rapidly gained traction in the past several decades as a less expensive (the cost of performing genomic analyses on blood samples is at least an order of magnitude less than on tissue samples), less invasive (requiring only a simple blood draw) alternative source of information about tumors.1
As researchers have refined the ability to exploit liquid biopsies, commercial interest has been piqued. More than 35 companies within the United States alone are developing liquid biopsies, and it’s easy to see why with a market projected to be in the many billions of dollars.2
Seeking out tumor clues in the blood
Liquid biopsies consist of a 10-15 mL blood sample drawn into a tube that contains an anticoagulant and it can contain several different types of tumor-associated material. Thus far, two components – circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA) – have formed the cornerstone of liquid biopsies. At present, it is not clear whether these components are released randomly, as a by-product of tumor cell death or if they are released as part of a specific biologic process, such as for the colonization of metastatic sites. It reality, it may be a little of both, and active dissemination may be particularly relevant for CTCs, among which are postulated to be a population of cancer stem cells that can initiate distant metastases.3,4
The discovery of CTCs dates back to the 1860s, when cells that were morphologically identical to the tumor were identified in the blood of a patient with metastatic cancer. Their potential significance was not fully realized until a few decades ago, when they were found to exist from early on in the course of disease.3,4
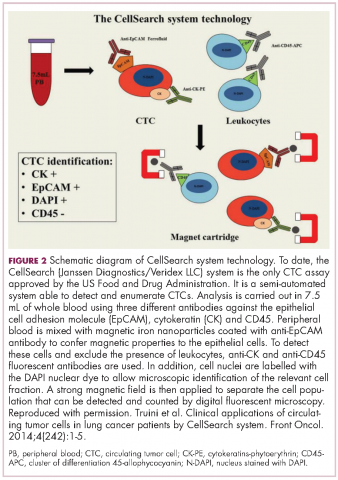
CTCs, which can be either single cells or clusters of cells known as microemboli, have a short half-life in the bloodstream – less than 2 ½ hours – and are also extremely rare (1 mL of blood contains 1-10 CTCs) against a background of many millions of normal cells. Thus the detection and isolation of CTCs presents a significant challenge. More than 40 different platforms are being developed for the isolation and enrichment of CTCs. For the most part, these use a method called positive selection to pick out CTCs.1,3,4
Positive selection exploits the biological or physical properties that are specific to CTCs and absent in normal cells, for example, the presence of a specific tumor-associated antigen on their surface or differences in size, density or electric charge. The limitations of this method are that, not only do you need to know something about CTCs to begin to understand what makes them truly unique and ensure only isolation of CTCs, but their phenotype is also thought to be continually changing.1,3,4
In recent years, the focus has shifted toward technologies that use negative depletion, meaning that they target the other types of cells in the blood sample and filter those away until only the CTCs are left behind. The most advanced are devices that use microfluidic technology to sort the cells, such as the CTC-iChip system being developed by researchers at Massachusetts General Hospital in Boston.5
ctDNA consists of small fragments of nucleic acids that are not contained within a cell or associated with cell fragments and is thought to be present in 50%-90% of patients, depending on the type of cancer they have. ctDNA has a similarly short half-life in the circulation to CTCs and, like CTCs, ctDNA is present at very low levels in the bloodstream. Although levels of ctDNA have been shown to increase with increasing tumor burden, it is still often obscured by the presence of other cell-free DNA derived from non-tumor cells.
ctDNA can be distinguished from other cell-free DNA by the presence of somatic mutations and a number of highly sensitive methods have been developed to detect them, including the amplification-refractory mutation system (ARMS); digital polymerase chain reaction; and the beads, emulsification, amplification, and magnetics (BEAMing) system. Next-generation sequencing technologies, including tagged-amplicon deep sequencing (TAm-Seq), the Safe-Sequencing System (Safe-SeqS), and cancer personalized profiling by deep sequencing (CAPP-seq), can also be used and the race for ever more sensitive analytical tools is ongoing.1,3,4,6
Applying liquid biopsies now and in the future
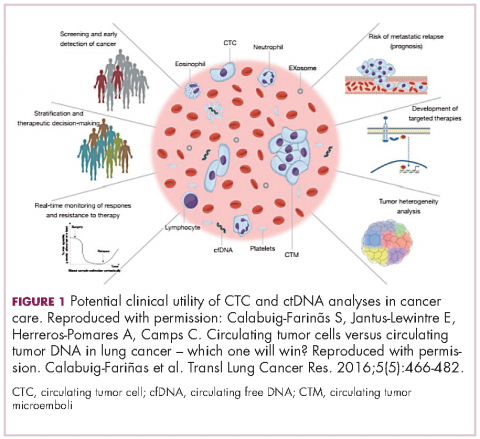
There are a plethora of potential applications for liquid biopsies3,7 (Figure 1), and probably the most exciting among them is the potential for screening for and early detection of cancer. The fact that ctDNA and CTCs have both been shown to be present from the earliest stages of disease has sparked interest in the possibility of developing simple blood tests to identify tumors before they become detectable by other methods and at a point at which they may be curable.
Given that both are present at such low levels within the circulation and are particularly sparse at earlier stages of disease, current technologies may lack the specificity and sensitivity for this application at present. However, numerous clinical trials are ongoing.
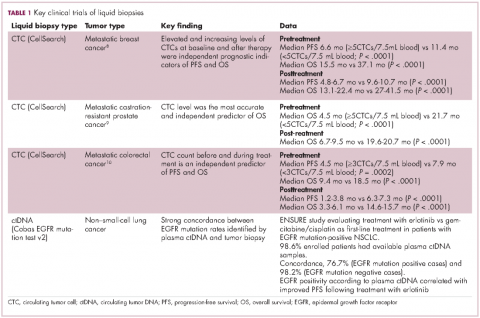
For CTCs, simple enumeration has been the most extensively investigated application to date. Numerous studies have shown that the number of CTCs in the bloodstream has prognostic significance in various different tumor types. Three such studies led to the first regulatory approval for a CTC detection system (Table 1 and Table 2).8-10
One area in which liquid biopsies could really come into their own is in providing more real-time analysis of tumors. This is something that has proven particularly challenging with tissue biopsies because repeating these invasive procedures is problematic. But the ease of repeat blood draws means that serial liquid biopsies could be performed and might offer the possibility of monitoring disease progression and evolution over the course of disease and particularly in response to treatment.
Indeed, studies have shown that in addition to baseline CTC counts, changes in CTC number during treatment are also prognostic. There was improved survival among patients whose CTC counts decreased below a threshold value during treatment and vice versa. This is also an approved use for CellSearch though at present it is not widely clinically implemented.12
Clinical utility remains elusive
The ultimate goal would be for liquid biopsies to have an impact on treatment decisions, allowing oncologists to change management strategy based on predicted sensitivity or resistance to therapy, so-called clinical utility. Thus far, clinical utility has proved elusive, though liquid biopsies using ctDNA to evaluate tumor genotype have come closest.
The Cobas EGFR Mutation Test v2 recently became the first ctDNA-based liquid biopsy to receive regulatory approval. It was approved as a companion diagnostic to identify patients with advanced non–small-cell lung cancer (NSCLC) who have specific mutations in the epidermal growth factor receptor (EGFR) gene and are therefore eligible for treatment with the EGFR inhibitor erlotinib.13
Approval was based on comparison of EGFR mutation identification rates using plasma ctDNA samples and tumor tissue samples from patients enrolled in the phase 3 ENSURE trial, which compared the efficacy of erlotinib with chemotherapy as first-line therapy in patients with advanced NSCLC. Of the 217 patients enrolled in the trial, 98.6% of patients had both tumor biopsy and plasma ctDNA samples available for testing. Concordance between the two types of biopsy in identifying patients with EGFR mutations was high and patients with EGFR positivity according to liquid biopsy results demonstrated improved progression-free survival when treated with erlotinib.14
The results of a large-scale genomic analysis of various different types of tumors using ctDNA were also recently presented at the 2016 American Society of Clinical Oncology meeting. Blood samples from more than 15,000 patients with 50 different tumor types, including advanced lung cancer (37%), breast cancer (14%), and CRC (10%), were collected and compared with either available tumor biopsy samples from the same cases (n = 398) or, in the majority of cases, with The Cancer Genome Atlas database, which uses tumor biopsies to perform genome-wide sequencing studies. Both types of biopsy revealed very similar mutation patterns when the Guardant360 next-generation sequencing test, which targets 70 genes, was applied. In particular, when EGFR, BRAF, KRAS, ALK, RET, and ROS1 mutations were identified by tumor tissue biopsy, the same mutations were reported in 94%-100% of plasma samples.15
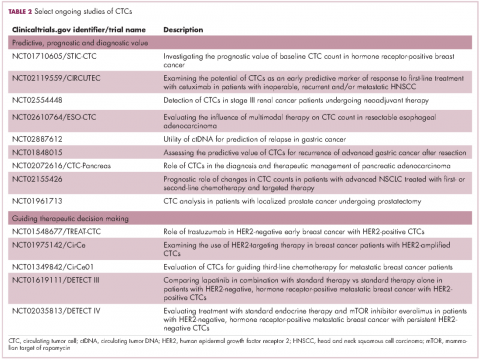
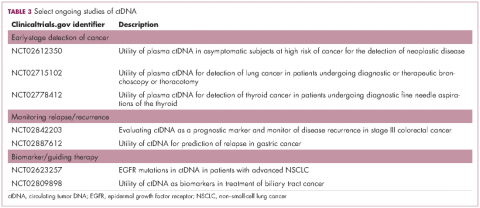
Studies of the clinical utility of ctDNA and CTCs are among ongoing clinical trials of liquid biopsies (Tables 2 and 3). The potential for using CTCs to guide treatment decisions has become particularly relevant in breast cancer in light of results showing that patients with primary tumors that are negative for human epidermal growth factor receptor 2 (HER2) amplification, an important biomarker in breast cancer, may have CTCs that are HER2-positive, in up to 30% of cases. These patients may therefore still benefit from HER2-targeted therapy.16
The DETECT studies are the first phase 3 trials in which treatment decisions are being based on the phenotypic characteristics of CTCs. DETECT III (NCT01619111) is comparing lapatinib in combination with standard therapy with standard therapy alone in patients with HER2-negative metastatic breast cancer who have HER2-positive CTCs, whereas DETECT IV (NCT02035813) is enrolling patients with HER2-negative, hormone receptor-positive metastatic breast cancer and persistent HER2-negative CTCs to receive standard endocrine therapy and the mammalian target of rapamycin inhibitor everolimus.
Other targets and sources for liquid biopsy
Another approach to liquid biopsies that is also beginning to take off is to collect tumor-derived exosomes from the bloodstream. Exosomes are tiny, fluid-filled, membrane-bound sacks that bud off from the surface of a cell to expel waste or to transport cargo from one cell to another. DNA, RNA, and protein can be extracted from tumor-derived exosomes and could also serve as molecular biomarkers relating to the cancer cells from which they came.6,7
Exosome Diagnostics is bringing the first exosome-based diagnostic tests to the market and recently teamed up with Amgen for the development of these liquid biopsies.17 In January 2016, they launched ExoDx Lung (ALK), for detection of EML4-ALK gene fusions in patients with NSCLC, using a proprietary platform for the isolation of RNA from exosomes. Data that was presented at several different conferences in 2015 demonstrated a sensitivity of 88% and specificity of 100% for this diagnostic when compared with tissue ALK status in NSCLC patients receiving a second-generation ALK inhibitor following progression on prior ALK inhibitor therapy.18
In September, they subsequently announced the launch of a test that analyses genetic information from exosomes collected from a urine sample taken from prostate cancer patients. Using a 3-gene signature, in combination with a proprietary algorithm, this diagnostic generates a score assessing a prostate cancer patient’s risk for higher grade, more aggressive disease. It is designed to complement the prostate-specific antigen score and has demonstrated accuracy in ruling out the presence of high-grade cancer before an initial biopsy in more than 1,
1. Lennon NK, Adalsteinsson VA, Gabriel SB. Technological considerations for genome-guided diagnosis and management of cancer. Genome Med. 2016;8:112.
2. MIT Technology Review website. Liquid biopsy: fast DNA-sequencing machines are leading to simple blood tests for cancer. https://www.technologyreview.com/s/534991/liquid-biopsy/. Published 2015. Accessed December 19, 2016.
3. Alix-Panabières C and Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6(5):479-491.
4. Calabuig-Farinãs S, Jantus-Lewintre E, Herreros-Pomares A, Camps C. Circulating tumor cells versus circulating tumor DNA in lung cancer – which one will win? Transl Lung Cancer Res. 2016;5(5):466-482.
5. Karabacak, NM, Spuhler PS, Fachin F, et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc. 2014;9:694-710.
6. Buder A, Tomuta C, and Filipits M. The potential of liquid biopsies. Curr Opin Oncol. 2016;28:130-134.
7. Hofman P, Popper HH. Pathologists and liquid biopsies: to be or not to be? Virchows Arch. 2016;469:601-609.
8. Bidard FC, Peeters DJ, Fehm T, et al. Clinical validity of circulating tumor cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 2014;15(4):406-414.
9. de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14(19):6302-6309.
10. Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(19):3213-3221.
11. CellSearch Web site. What is the CELLSEARCH® System? https://www.cellsearchctc.com/product-systems-overview/cellsearch-system-overview. Last updated December 5th, 2016. Accessed online December 19th, 2016.
12. CellSearch Web site [advertisement]. https://www.cellsearchctc.com/clinical-applications/clinical-applications-overview. Last updated December 5, 2016. Accessed December 19, 2016.
13. US Food and Drug Administration. cobas EGFR Mutation Test v2 – P150047. http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/DeviceApprovalsandClearances/Recently-ApprovedDevices/ucm519922.htm. Last updated September 9, 2016. Accessed December 19, 2016.
14. Wu YL, Zhou C, Liam CK, et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small cell lung cancer: analyses from the phase III, randomized, open-label, ENSURE study. Ann Oncol. 2015;26(9):1883-1889.
15. Zill OA, Mortimer S, Banks KC, et al. Somatic genomic landscape of over 15,000 patients with advanced-stage cancer from clinical next-generation sequencing analysis of circulating tumor DNA. J Clin Oncol. 2016;34(suppl;abstr LBA11501).
16. Jordan NV, Bardia A, Wittner BS, et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature. 2016;537:102-106.
17. Exosome Diagnostics. Exosome diagnostics enters agreement with Amgen. http://www.exosomedx.com/news-events/press-releases/exosome-diagnostics-enters-agreement-amgen. Published October 3, 2016. Accessed December 19, 2016.
18. Brinkman K, Emenegger J, Tannous B, et al. Exosomal RNA-based liquid biopsy detection of EML4-ALK in plasma from NSCLC patients [2015 World Conference on Lung Cancer, Denver, CO; abstract 2591]. http://library.iaslc.org/search-speaker?search_speaker=30493. Accessed January 6, 2017.
19. Exosome Diagnostics website. Prostate cancer. http://www.exosomedx.com/prostate-cancer-0. Last updated 2017. Accessed online December 19, 2016.
Pathologic and, increasingly, molecular analysis of tumor tissue biopsies is the gold standard in initial diagnosis of cancer, but liquid biopsies, which analyze tumor-derived material circulating in the bloodstream are gaining traction. Here, we discuss the current state of development of this complementary and potentially alternative approach to tumor analysis.
Liquid biopsy gaining traction
Biopsies enable oncologists to gather information about a potential or established tumor, including confirmation of the presence of cancerous tissue and determination of its histological characteristics, such as tumor grade and stage, as well as its molecular features, such as the presence of certain gene mutations. Ultimately, this information can be put to use in determining the most appropriate course of treatment.
The current gold standard is a tissue biopsy that typically involves an invasive procedure to permit the collection of a piece of tumor tissue. Yet, tissue biopsies are not always feasible because of the location of the tumor or the poor performance status of many patients with advanced disease. They also provide only a snapshot of the disease at the time at which they were taken and don’t necessarily reflect the genetic heterogeneity or evolution of a tumor over time.
The detection of components that are derived from the tumor circulating in the blood of cancer patients had fueled the idea of blood-based diagnostics in oncology – so-called liquid biopsies. These have rapidly gained traction in the past several decades as a less expensive (the cost of performing genomic analyses on blood samples is at least an order of magnitude less than on tissue samples), less invasive (requiring only a simple blood draw) alternative source of information about tumors.1
As researchers have refined the ability to exploit liquid biopsies, commercial interest has been piqued. More than 35 companies within the United States alone are developing liquid biopsies, and it’s easy to see why with a market projected to be in the many billions of dollars.2
Seeking out tumor clues in the blood
Liquid biopsies consist of a 10-15 mL blood sample drawn into a tube that contains an anticoagulant and it can contain several different types of tumor-associated material. Thus far, two components – circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA) – have formed the cornerstone of liquid biopsies. At present, it is not clear whether these components are released randomly, as a by-product of tumor cell death or if they are released as part of a specific biologic process, such as for the colonization of metastatic sites. It reality, it may be a little of both, and active dissemination may be particularly relevant for CTCs, among which are postulated to be a population of cancer stem cells that can initiate distant metastases.3,4
The discovery of CTCs dates back to the 1860s, when cells that were morphologically identical to the tumor were identified in the blood of a patient with metastatic cancer. Their potential significance was not fully realized until a few decades ago, when they were found to exist from early on in the course of disease.3,4
CTCs, which can be either single cells or clusters of cells known as microemboli, have a short half-life in the bloodstream – less than 2 ½ hours – and are also extremely rare (1 mL of blood contains 1-10 CTCs) against a background of many millions of normal cells. Thus the detection and isolation of CTCs presents a significant challenge. More than 40 different platforms are being developed for the isolation and enrichment of CTCs. For the most part, these use a method called positive selection to pick out CTCs.1,3,4
Positive selection exploits the biological or physical properties that are specific to CTCs and absent in normal cells, for example, the presence of a specific tumor-associated antigen on their surface or differences in size, density or electric charge. The limitations of this method are that, not only do you need to know something about CTCs to begin to understand what makes them truly unique and ensure only isolation of CTCs, but their phenotype is also thought to be continually changing.1,3,4
In recent years, the focus has shifted toward technologies that use negative depletion, meaning that they target the other types of cells in the blood sample and filter those away until only the CTCs are left behind. The most advanced are devices that use microfluidic technology to sort the cells, such as the CTC-iChip system being developed by researchers at Massachusetts General Hospital in Boston.5
ctDNA consists of small fragments of nucleic acids that are not contained within a cell or associated with cell fragments and is thought to be present in 50%-90% of patients, depending on the type of cancer they have. ctDNA has a similarly short half-life in the circulation to CTCs and, like CTCs, ctDNA is present at very low levels in the bloodstream. Although levels of ctDNA have been shown to increase with increasing tumor burden, it is still often obscured by the presence of other cell-free DNA derived from non-tumor cells.
ctDNA can be distinguished from other cell-free DNA by the presence of somatic mutations and a number of highly sensitive methods have been developed to detect them, including the amplification-refractory mutation system (ARMS); digital polymerase chain reaction; and the beads, emulsification, amplification, and magnetics (BEAMing) system. Next-generation sequencing technologies, including tagged-amplicon deep sequencing (TAm-Seq), the Safe-Sequencing System (Safe-SeqS), and cancer personalized profiling by deep sequencing (CAPP-seq), can also be used and the race for ever more sensitive analytical tools is ongoing.1,3,4,6
Applying liquid biopsies now and in the future
There are a plethora of potential applications for liquid biopsies3,7 (Figure 1), and probably the most exciting among them is the potential for screening for and early detection of cancer. The fact that ctDNA and CTCs have both been shown to be present from the earliest stages of disease has sparked interest in the possibility of developing simple blood tests to identify tumors before they become detectable by other methods and at a point at which they may be curable.
Given that both are present at such low levels within the circulation and are particularly sparse at earlier stages of disease, current technologies may lack the specificity and sensitivity for this application at present. However, numerous clinical trials are ongoing.
For CTCs, simple enumeration has been the most extensively investigated application to date. Numerous studies have shown that the number of CTCs in the bloodstream has prognostic significance in various different tumor types. Three such studies led to the first regulatory approval for a CTC detection system (Table 1 and Table 2).8-10
One area in which liquid biopsies could really come into their own is in providing more real-time analysis of tumors. This is something that has proven particularly challenging with tissue biopsies because repeating these invasive procedures is problematic. But the ease of repeat blood draws means that serial liquid biopsies could be performed and might offer the possibility of monitoring disease progression and evolution over the course of disease and particularly in response to treatment.
Indeed, studies have shown that in addition to baseline CTC counts, changes in CTC number during treatment are also prognostic. There was improved survival among patients whose CTC counts decreased below a threshold value during treatment and vice versa. This is also an approved use for CellSearch though at present it is not widely clinically implemented.12
Clinical utility remains elusive
The ultimate goal would be for liquid biopsies to have an impact on treatment decisions, allowing oncologists to change management strategy based on predicted sensitivity or resistance to therapy, so-called clinical utility. Thus far, clinical utility has proved elusive, though liquid biopsies using ctDNA to evaluate tumor genotype have come closest.
The Cobas EGFR Mutation Test v2 recently became the first ctDNA-based liquid biopsy to receive regulatory approval. It was approved as a companion diagnostic to identify patients with advanced non–small-cell lung cancer (NSCLC) who have specific mutations in the epidermal growth factor receptor (EGFR) gene and are therefore eligible for treatment with the EGFR inhibitor erlotinib.13
Approval was based on comparison of EGFR mutation identification rates using plasma ctDNA samples and tumor tissue samples from patients enrolled in the phase 3 ENSURE trial, which compared the efficacy of erlotinib with chemotherapy as first-line therapy in patients with advanced NSCLC. Of the 217 patients enrolled in the trial, 98.6% of patients had both tumor biopsy and plasma ctDNA samples available for testing. Concordance between the two types of biopsy in identifying patients with EGFR mutations was high and patients with EGFR positivity according to liquid biopsy results demonstrated improved progression-free survival when treated with erlotinib.14
The results of a large-scale genomic analysis of various different types of tumors using ctDNA were also recently presented at the 2016 American Society of Clinical Oncology meeting. Blood samples from more than 15,000 patients with 50 different tumor types, including advanced lung cancer (37%), breast cancer (14%), and CRC (10%), were collected and compared with either available tumor biopsy samples from the same cases (n = 398) or, in the majority of cases, with The Cancer Genome Atlas database, which uses tumor biopsies to perform genome-wide sequencing studies. Both types of biopsy revealed very similar mutation patterns when the Guardant360 next-generation sequencing test, which targets 70 genes, was applied. In particular, when EGFR, BRAF, KRAS, ALK, RET, and ROS1 mutations were identified by tumor tissue biopsy, the same mutations were reported in 94%-100% of plasma samples.15
Studies of the clinical utility of ctDNA and CTCs are among ongoing clinical trials of liquid biopsies (Tables 2 and 3). The potential for using CTCs to guide treatment decisions has become particularly relevant in breast cancer in light of results showing that patients with primary tumors that are negative for human epidermal growth factor receptor 2 (HER2) amplification, an important biomarker in breast cancer, may have CTCs that are HER2-positive, in up to 30% of cases. These patients may therefore still benefit from HER2-targeted therapy.16
The DETECT studies are the first phase 3 trials in which treatment decisions are being based on the phenotypic characteristics of CTCs. DETECT III (NCT01619111) is comparing lapatinib in combination with standard therapy with standard therapy alone in patients with HER2-negative metastatic breast cancer who have HER2-positive CTCs, whereas DETECT IV (NCT02035813) is enrolling patients with HER2-negative, hormone receptor-positive metastatic breast cancer and persistent HER2-negative CTCs to receive standard endocrine therapy and the mammalian target of rapamycin inhibitor everolimus.
Other targets and sources for liquid biopsy
Another approach to liquid biopsies that is also beginning to take off is to collect tumor-derived exosomes from the bloodstream. Exosomes are tiny, fluid-filled, membrane-bound sacks that bud off from the surface of a cell to expel waste or to transport cargo from one cell to another. DNA, RNA, and protein can be extracted from tumor-derived exosomes and could also serve as molecular biomarkers relating to the cancer cells from which they came.6,7
Exosome Diagnostics is bringing the first exosome-based diagnostic tests to the market and recently teamed up with Amgen for the development of these liquid biopsies.17 In January 2016, they launched ExoDx Lung (ALK), for detection of EML4-ALK gene fusions in patients with NSCLC, using a proprietary platform for the isolation of RNA from exosomes. Data that was presented at several different conferences in 2015 demonstrated a sensitivity of 88% and specificity of 100% for this diagnostic when compared with tissue ALK status in NSCLC patients receiving a second-generation ALK inhibitor following progression on prior ALK inhibitor therapy.18
In September, they subsequently announced the launch of a test that analyses genetic information from exosomes collected from a urine sample taken from prostate cancer patients. Using a 3-gene signature, in combination with a proprietary algorithm, this diagnostic generates a score assessing a prostate cancer patient’s risk for higher grade, more aggressive disease. It is designed to complement the prostate-specific antigen score and has demonstrated accuracy in ruling out the presence of high-grade cancer before an initial biopsy in more than 1,
Pathologic and, increasingly, molecular analysis of tumor tissue biopsies is the gold standard in initial diagnosis of cancer, but liquid biopsies, which analyze tumor-derived material circulating in the bloodstream are gaining traction. Here, we discuss the current state of development of this complementary and potentially alternative approach to tumor analysis.
Liquid biopsy gaining traction
Biopsies enable oncologists to gather information about a potential or established tumor, including confirmation of the presence of cancerous tissue and determination of its histological characteristics, such as tumor grade and stage, as well as its molecular features, such as the presence of certain gene mutations. Ultimately, this information can be put to use in determining the most appropriate course of treatment.
The current gold standard is a tissue biopsy that typically involves an invasive procedure to permit the collection of a piece of tumor tissue. Yet, tissue biopsies are not always feasible because of the location of the tumor or the poor performance status of many patients with advanced disease. They also provide only a snapshot of the disease at the time at which they were taken and don’t necessarily reflect the genetic heterogeneity or evolution of a tumor over time.
The detection of components that are derived from the tumor circulating in the blood of cancer patients had fueled the idea of blood-based diagnostics in oncology – so-called liquid biopsies. These have rapidly gained traction in the past several decades as a less expensive (the cost of performing genomic analyses on blood samples is at least an order of magnitude less than on tissue samples), less invasive (requiring only a simple blood draw) alternative source of information about tumors.1
As researchers have refined the ability to exploit liquid biopsies, commercial interest has been piqued. More than 35 companies within the United States alone are developing liquid biopsies, and it’s easy to see why with a market projected to be in the many billions of dollars.2
Seeking out tumor clues in the blood
Liquid biopsies consist of a 10-15 mL blood sample drawn into a tube that contains an anticoagulant and it can contain several different types of tumor-associated material. Thus far, two components – circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA) – have formed the cornerstone of liquid biopsies. At present, it is not clear whether these components are released randomly, as a by-product of tumor cell death or if they are released as part of a specific biologic process, such as for the colonization of metastatic sites. It reality, it may be a little of both, and active dissemination may be particularly relevant for CTCs, among which are postulated to be a population of cancer stem cells that can initiate distant metastases.3,4
The discovery of CTCs dates back to the 1860s, when cells that were morphologically identical to the tumor were identified in the blood of a patient with metastatic cancer. Their potential significance was not fully realized until a few decades ago, when they were found to exist from early on in the course of disease.3,4
CTCs, which can be either single cells or clusters of cells known as microemboli, have a short half-life in the bloodstream – less than 2 ½ hours – and are also extremely rare (1 mL of blood contains 1-10 CTCs) against a background of many millions of normal cells. Thus the detection and isolation of CTCs presents a significant challenge. More than 40 different platforms are being developed for the isolation and enrichment of CTCs. For the most part, these use a method called positive selection to pick out CTCs.1,3,4
Positive selection exploits the biological or physical properties that are specific to CTCs and absent in normal cells, for example, the presence of a specific tumor-associated antigen on their surface or differences in size, density or electric charge. The limitations of this method are that, not only do you need to know something about CTCs to begin to understand what makes them truly unique and ensure only isolation of CTCs, but their phenotype is also thought to be continually changing.1,3,4
In recent years, the focus has shifted toward technologies that use negative depletion, meaning that they target the other types of cells in the blood sample and filter those away until only the CTCs are left behind. The most advanced are devices that use microfluidic technology to sort the cells, such as the CTC-iChip system being developed by researchers at Massachusetts General Hospital in Boston.5
ctDNA consists of small fragments of nucleic acids that are not contained within a cell or associated with cell fragments and is thought to be present in 50%-90% of patients, depending on the type of cancer they have. ctDNA has a similarly short half-life in the circulation to CTCs and, like CTCs, ctDNA is present at very low levels in the bloodstream. Although levels of ctDNA have been shown to increase with increasing tumor burden, it is still often obscured by the presence of other cell-free DNA derived from non-tumor cells.
ctDNA can be distinguished from other cell-free DNA by the presence of somatic mutations and a number of highly sensitive methods have been developed to detect them, including the amplification-refractory mutation system (ARMS); digital polymerase chain reaction; and the beads, emulsification, amplification, and magnetics (BEAMing) system. Next-generation sequencing technologies, including tagged-amplicon deep sequencing (TAm-Seq), the Safe-Sequencing System (Safe-SeqS), and cancer personalized profiling by deep sequencing (CAPP-seq), can also be used and the race for ever more sensitive analytical tools is ongoing.1,3,4,6
Applying liquid biopsies now and in the future
There are a plethora of potential applications for liquid biopsies3,7 (Figure 1), and probably the most exciting among them is the potential for screening for and early detection of cancer. The fact that ctDNA and CTCs have both been shown to be present from the earliest stages of disease has sparked interest in the possibility of developing simple blood tests to identify tumors before they become detectable by other methods and at a point at which they may be curable.
Given that both are present at such low levels within the circulation and are particularly sparse at earlier stages of disease, current technologies may lack the specificity and sensitivity for this application at present. However, numerous clinical trials are ongoing.
For CTCs, simple enumeration has been the most extensively investigated application to date. Numerous studies have shown that the number of CTCs in the bloodstream has prognostic significance in various different tumor types. Three such studies led to the first regulatory approval for a CTC detection system (Table 1 and Table 2).8-10
One area in which liquid biopsies could really come into their own is in providing more real-time analysis of tumors. This is something that has proven particularly challenging with tissue biopsies because repeating these invasive procedures is problematic. But the ease of repeat blood draws means that serial liquid biopsies could be performed and might offer the possibility of monitoring disease progression and evolution over the course of disease and particularly in response to treatment.
Indeed, studies have shown that in addition to baseline CTC counts, changes in CTC number during treatment are also prognostic. There was improved survival among patients whose CTC counts decreased below a threshold value during treatment and vice versa. This is also an approved use for CellSearch though at present it is not widely clinically implemented.12
Clinical utility remains elusive
The ultimate goal would be for liquid biopsies to have an impact on treatment decisions, allowing oncologists to change management strategy based on predicted sensitivity or resistance to therapy, so-called clinical utility. Thus far, clinical utility has proved elusive, though liquid biopsies using ctDNA to evaluate tumor genotype have come closest.
The Cobas EGFR Mutation Test v2 recently became the first ctDNA-based liquid biopsy to receive regulatory approval. It was approved as a companion diagnostic to identify patients with advanced non–small-cell lung cancer (NSCLC) who have specific mutations in the epidermal growth factor receptor (EGFR) gene and are therefore eligible for treatment with the EGFR inhibitor erlotinib.13
Approval was based on comparison of EGFR mutation identification rates using plasma ctDNA samples and tumor tissue samples from patients enrolled in the phase 3 ENSURE trial, which compared the efficacy of erlotinib with chemotherapy as first-line therapy in patients with advanced NSCLC. Of the 217 patients enrolled in the trial, 98.6% of patients had both tumor biopsy and plasma ctDNA samples available for testing. Concordance between the two types of biopsy in identifying patients with EGFR mutations was high and patients with EGFR positivity according to liquid biopsy results demonstrated improved progression-free survival when treated with erlotinib.14
The results of a large-scale genomic analysis of various different types of tumors using ctDNA were also recently presented at the 2016 American Society of Clinical Oncology meeting. Blood samples from more than 15,000 patients with 50 different tumor types, including advanced lung cancer (37%), breast cancer (14%), and CRC (10%), were collected and compared with either available tumor biopsy samples from the same cases (n = 398) or, in the majority of cases, with The Cancer Genome Atlas database, which uses tumor biopsies to perform genome-wide sequencing studies. Both types of biopsy revealed very similar mutation patterns when the Guardant360 next-generation sequencing test, which targets 70 genes, was applied. In particular, when EGFR, BRAF, KRAS, ALK, RET, and ROS1 mutations were identified by tumor tissue biopsy, the same mutations were reported in 94%-100% of plasma samples.15
Studies of the clinical utility of ctDNA and CTCs are among ongoing clinical trials of liquid biopsies (Tables 2 and 3). The potential for using CTCs to guide treatment decisions has become particularly relevant in breast cancer in light of results showing that patients with primary tumors that are negative for human epidermal growth factor receptor 2 (HER2) amplification, an important biomarker in breast cancer, may have CTCs that are HER2-positive, in up to 30% of cases. These patients may therefore still benefit from HER2-targeted therapy.16
The DETECT studies are the first phase 3 trials in which treatment decisions are being based on the phenotypic characteristics of CTCs. DETECT III (NCT01619111) is comparing lapatinib in combination with standard therapy with standard therapy alone in patients with HER2-negative metastatic breast cancer who have HER2-positive CTCs, whereas DETECT IV (NCT02035813) is enrolling patients with HER2-negative, hormone receptor-positive metastatic breast cancer and persistent HER2-negative CTCs to receive standard endocrine therapy and the mammalian target of rapamycin inhibitor everolimus.
Other targets and sources for liquid biopsy
Another approach to liquid biopsies that is also beginning to take off is to collect tumor-derived exosomes from the bloodstream. Exosomes are tiny, fluid-filled, membrane-bound sacks that bud off from the surface of a cell to expel waste or to transport cargo from one cell to another. DNA, RNA, and protein can be extracted from tumor-derived exosomes and could also serve as molecular biomarkers relating to the cancer cells from which they came.6,7
Exosome Diagnostics is bringing the first exosome-based diagnostic tests to the market and recently teamed up with Amgen for the development of these liquid biopsies.17 In January 2016, they launched ExoDx Lung (ALK), for detection of EML4-ALK gene fusions in patients with NSCLC, using a proprietary platform for the isolation of RNA from exosomes. Data that was presented at several different conferences in 2015 demonstrated a sensitivity of 88% and specificity of 100% for this diagnostic when compared with tissue ALK status in NSCLC patients receiving a second-generation ALK inhibitor following progression on prior ALK inhibitor therapy.18
In September, they subsequently announced the launch of a test that analyses genetic information from exosomes collected from a urine sample taken from prostate cancer patients. Using a 3-gene signature, in combination with a proprietary algorithm, this diagnostic generates a score assessing a prostate cancer patient’s risk for higher grade, more aggressive disease. It is designed to complement the prostate-specific antigen score and has demonstrated accuracy in ruling out the presence of high-grade cancer before an initial biopsy in more than 1,
1. Lennon NK, Adalsteinsson VA, Gabriel SB. Technological considerations for genome-guided diagnosis and management of cancer. Genome Med. 2016;8:112.
2. MIT Technology Review website. Liquid biopsy: fast DNA-sequencing machines are leading to simple blood tests for cancer. https://www.technologyreview.com/s/534991/liquid-biopsy/. Published 2015. Accessed December 19, 2016.
3. Alix-Panabières C and Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6(5):479-491.
4. Calabuig-Farinãs S, Jantus-Lewintre E, Herreros-Pomares A, Camps C. Circulating tumor cells versus circulating tumor DNA in lung cancer – which one will win? Transl Lung Cancer Res. 2016;5(5):466-482.
5. Karabacak, NM, Spuhler PS, Fachin F, et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc. 2014;9:694-710.
6. Buder A, Tomuta C, and Filipits M. The potential of liquid biopsies. Curr Opin Oncol. 2016;28:130-134.
7. Hofman P, Popper HH. Pathologists and liquid biopsies: to be or not to be? Virchows Arch. 2016;469:601-609.
8. Bidard FC, Peeters DJ, Fehm T, et al. Clinical validity of circulating tumor cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 2014;15(4):406-414.
9. de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14(19):6302-6309.
10. Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(19):3213-3221.
11. CellSearch Web site. What is the CELLSEARCH® System? https://www.cellsearchctc.com/product-systems-overview/cellsearch-system-overview. Last updated December 5th, 2016. Accessed online December 19th, 2016.
12. CellSearch Web site [advertisement]. https://www.cellsearchctc.com/clinical-applications/clinical-applications-overview. Last updated December 5, 2016. Accessed December 19, 2016.
13. US Food and Drug Administration. cobas EGFR Mutation Test v2 – P150047. http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/DeviceApprovalsandClearances/Recently-ApprovedDevices/ucm519922.htm. Last updated September 9, 2016. Accessed December 19, 2016.
14. Wu YL, Zhou C, Liam CK, et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small cell lung cancer: analyses from the phase III, randomized, open-label, ENSURE study. Ann Oncol. 2015;26(9):1883-1889.
15. Zill OA, Mortimer S, Banks KC, et al. Somatic genomic landscape of over 15,000 patients with advanced-stage cancer from clinical next-generation sequencing analysis of circulating tumor DNA. J Clin Oncol. 2016;34(suppl;abstr LBA11501).
16. Jordan NV, Bardia A, Wittner BS, et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature. 2016;537:102-106.
17. Exosome Diagnostics. Exosome diagnostics enters agreement with Amgen. http://www.exosomedx.com/news-events/press-releases/exosome-diagnostics-enters-agreement-amgen. Published October 3, 2016. Accessed December 19, 2016.
18. Brinkman K, Emenegger J, Tannous B, et al. Exosomal RNA-based liquid biopsy detection of EML4-ALK in plasma from NSCLC patients [2015 World Conference on Lung Cancer, Denver, CO; abstract 2591]. http://library.iaslc.org/search-speaker?search_speaker=30493. Accessed January 6, 2017.
19. Exosome Diagnostics website. Prostate cancer. http://www.exosomedx.com/prostate-cancer-0. Last updated 2017. Accessed online December 19, 2016.
1. Lennon NK, Adalsteinsson VA, Gabriel SB. Technological considerations for genome-guided diagnosis and management of cancer. Genome Med. 2016;8:112.
2. MIT Technology Review website. Liquid biopsy: fast DNA-sequencing machines are leading to simple blood tests for cancer. https://www.technologyreview.com/s/534991/liquid-biopsy/. Published 2015. Accessed December 19, 2016.
3. Alix-Panabières C and Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6(5):479-491.
4. Calabuig-Farinãs S, Jantus-Lewintre E, Herreros-Pomares A, Camps C. Circulating tumor cells versus circulating tumor DNA in lung cancer – which one will win? Transl Lung Cancer Res. 2016;5(5):466-482.
5. Karabacak, NM, Spuhler PS, Fachin F, et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc. 2014;9:694-710.
6. Buder A, Tomuta C, and Filipits M. The potential of liquid biopsies. Curr Opin Oncol. 2016;28:130-134.
7. Hofman P, Popper HH. Pathologists and liquid biopsies: to be or not to be? Virchows Arch. 2016;469:601-609.
8. Bidard FC, Peeters DJ, Fehm T, et al. Clinical validity of circulating tumor cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 2014;15(4):406-414.
9. de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14(19):6302-6309.
10. Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(19):3213-3221.
11. CellSearch Web site. What is the CELLSEARCH® System? https://www.cellsearchctc.com/product-systems-overview/cellsearch-system-overview. Last updated December 5th, 2016. Accessed online December 19th, 2016.
12. CellSearch Web site [advertisement]. https://www.cellsearchctc.com/clinical-applications/clinical-applications-overview. Last updated December 5, 2016. Accessed December 19, 2016.
13. US Food and Drug Administration. cobas EGFR Mutation Test v2 – P150047. http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/DeviceApprovalsandClearances/Recently-ApprovedDevices/ucm519922.htm. Last updated September 9, 2016. Accessed December 19, 2016.
14. Wu YL, Zhou C, Liam CK, et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small cell lung cancer: analyses from the phase III, randomized, open-label, ENSURE study. Ann Oncol. 2015;26(9):1883-1889.
15. Zill OA, Mortimer S, Banks KC, et al. Somatic genomic landscape of over 15,000 patients with advanced-stage cancer from clinical next-generation sequencing analysis of circulating tumor DNA. J Clin Oncol. 2016;34(suppl;abstr LBA11501).
16. Jordan NV, Bardia A, Wittner BS, et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature. 2016;537:102-106.
17. Exosome Diagnostics. Exosome diagnostics enters agreement with Amgen. http://www.exosomedx.com/news-events/press-releases/exosome-diagnostics-enters-agreement-amgen. Published October 3, 2016. Accessed December 19, 2016.
18. Brinkman K, Emenegger J, Tannous B, et al. Exosomal RNA-based liquid biopsy detection of EML4-ALK in plasma from NSCLC patients [2015 World Conference on Lung Cancer, Denver, CO; abstract 2591]. http://library.iaslc.org/search-speaker?search_speaker=30493. Accessed January 6, 2017.
19. Exosome Diagnostics website. Prostate cancer. http://www.exosomedx.com/prostate-cancer-0. Last updated 2017. Accessed online December 19, 2016.