Whole exome sequencing’s role in diagnosing genetic causes of FGR with and without associated anomalies
Mone F, Mellis R, Gabriel H, et al. Should we offer prenatal exome sequencing for intrauterine growth restriction or short long bones? A systematic review and meta-analysis. Am J Obstet Gynecol. Published online October 7, 2022. doi:10.1016/j.ajog.2022.09.045
Multiple factors can play a role in FGR, including inherent maternal, placental, or fetal factors; the environment; and/or nutrition. However, prenatal diagnosis is an important consideration when exploring the underlying etiology for a growth-restricted fetus, especially in severe or early-onset cases. Many genetic conditions do not result in structural anomalies but can disrupt overall growth. Additionally, phenotyping in the prenatal period is limited and can miss more subtle physical differences that could point to a genetic cause.
When compared with karyotype, chromosomal microarray (CMA) has been shown to increase the diagnostic yield in cases of isolated early FGR by 5%,1,2 and the incidence of chromosomal abnormalities has been reported to be as high as 19% in this population. Let’s explore the data on exome sequencing for prenatal diagnosis in cases of isolated FGR.
Meta-analysis details
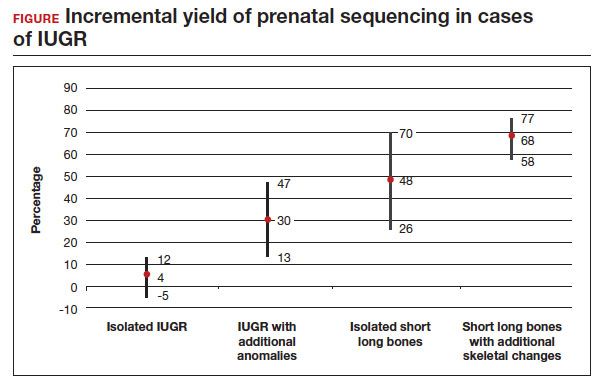
In this meta-analysis, the authors reviewed 19 cohort studies or case series that investigated the yield of prenatal sequencing in fetuses with intrauterine growth restriction (IUGR) or short long bones, both in association with and without additional anomalies. All cases had nondiagnostic cytogenetic results. Fetal DNA in most cases was obtained through amniocentesis. Variants classified as likely pathogenic and pathogenic were considered diagnostic. The authors then calculated the incremental yield of prenatal sequencing over cytogenetic studies as a pooled value, comparing the following groups:
- isolated FGR
- growth restriction with associated anomalies
- isolated short long bones
- short long bones with additional skeletal features.
Study outcomes
The total number of cases were as follows: isolated IUGR (n = 71), IUGR associated with additional anomalies (n = 45), isolated short long bones (n = 84), and short long bones associated with additional skeletal findings (n = 252). Causative pathogenic or likely pathogenic variants were identified in 224 (50%) cases. Apparent incremental yields with prenatal sequencing were as follows for the 4 groups (as illustrated in the FIGURE):
- 4% in isolated IUGR (95% confidence interval [CI], -5%–12%)
- 30% in IUGR with additional anomalies (95% CI, 13%–47%)
- 48% in isolated short long bones (95% CI, 26%–70%)
- 68% in short long bones with additional skeletal changes (95% CI, 58%–77%).
Overall, the authors concluded that prenatal sequencing does not improve prenatal diagnosis in cases of isolated IUGR. The majority of these cases were thought to be related to placental insufficiency.
Strengths and limitations
The main limitation of this study with regard to our discussion is the small study populationof isolated growth restriction. The authors indicate that the number of cases of isolated IUGR were too small to draw firm conclusions. Another limitation was the heterogeneity of the isolated FGR population, which was not limited to severe or early-onset cases. However, the authors did demonstrate that growth restriction in association with fetal anomalies has very high genetic yield rates with prenatal sequencing.
Not surprisingly, there is a high yield of diagnosing genetic conditions in pregnancies complicated by isolated or nonisolated short long bones or in cases of growth restriction with multisystem abnormalities. Based on the results of this study, the authors advise against sending for exome sequencing in cases of isolated growth restriction with coexisting evidence of placental insufficiency.
Continue to: Can whole exome sequencing diagnose genetic causes in cases of severe isolated FGR?...