The use of pharmacogenetic testing to help drive decisions for medication management of patients with psychiatric illnesses is growing. It’s becoming increasingly common for patients or the parents of pediatric patients to request pharmacogenetic testing or to bring the results of prior testing to their appointment. In these situations, patients may ask clinicians to consider the recommendations from these testing reports, which rarely provide guidance specific to pediatric patients. However, this can be difficult for clinicians who did not receive education in pharmacogenetics and may not be familiar with the evidence or options for pharmacogenetic testing. Many of the pharmacogenetic associations identified thus far have been discovered in adults, but studies in pediatric patients are relatively rare. This article reviews pharmacogenetic testing and the evidence supporting it, and describes implementation of routine pharmacogenetics testing at a children’s hospital.
CASE
Testing leads to dose adjustment, improvement
Ms. R, age 16, presents with treatment-resistant major depressive disorder that is characterized by a significant neurovegetative burden and prominent anhedonia, as well as intermittent suicidal ideation without intent or plan. She reportedly did not improve after multiple medication trials, including citalopram (maximum dose 30 mg/d, treatment duration 8 weeks, good compliance), sertraline (maximum dose 150 mg/d, treatment duration 10 weeks, good compliance), fluoxetine (maximum dose 40 mg/d, treatment duration 8 weeks, good compliance, mild improvement in neurovegetative symptoms and depressed mood), and duloxetine (maximum dose 90 mg/d, treatment duration 6 weeks, good compliance, mild benefit but intolerable nausea).
Augmentation strategies included risperidone, 1 mg/d at bedtime, but it failed to ameliorate her depressive symptoms. At the time of pharmacogenetic testing, she is taking aripiprazole, 2 mg/d at bedtime, and venlafaxine ER, 37.5 mg/d. Some benefit was noted, but her symptoms recrudesced within several weeks. Because both of these medications are metabolized by the cytochrome P450 (CYP) 2D6 enzyme, Ms. R is tested for CYP2D6 variants and is determined to be a CYP2D6 ultra-rapid metabolizer. Her venlafaxine ER is quickly titrated from 37.5 to 112.5 mg/d and aripiprazole is titrated from 2 to 10 mg/d. The patient’s anergia, amotivation, and mood improve.
_
Drug metabolism and genetic variants
It is common for patients with psychiatric disorders to receive trials of multiple psychotropic medications prior to identifying one that reduces symptom burden without producing intolerable adverse effects. Due to the high frequency of toxicity-related adverse effects (observed in 20% to 70% of patients),1 these medications are frequently initiated at low doses and titrated slowly until the patient either experiences an intolerable adverse effect or achieves symptomatic remission.1,2 The practice of slow titration at the start of treatment increases the risk of undertreatment in many patients, and may ultimately lead to a medication change due to the lack of response.
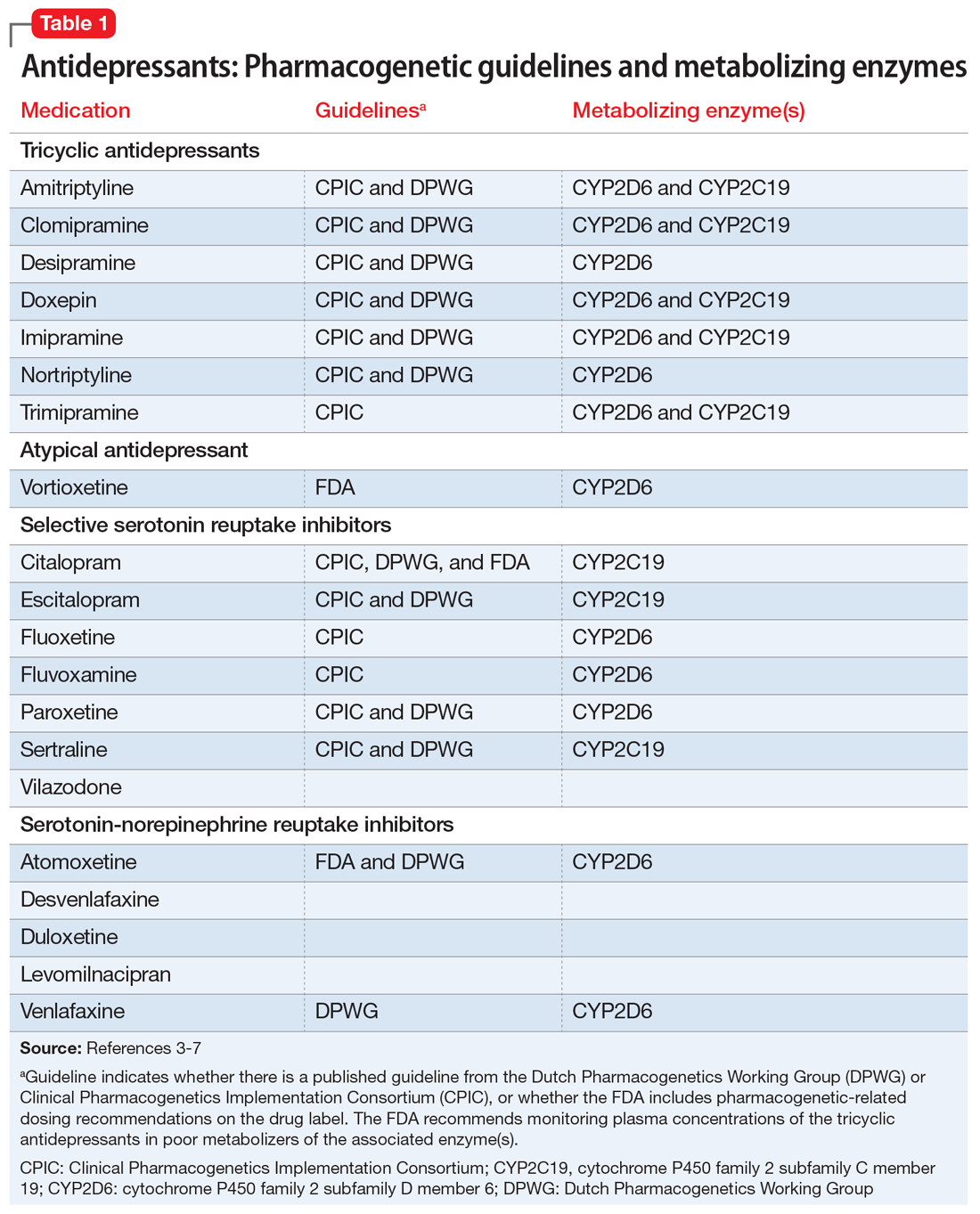
Many of the medications used to treat psychiatric illnesses are primarily metabolized by 2 CYP enzymes expressed in the liver, encoded by the CYP2D6 and CYP2C19 genes3 (Table 13-7 and Table 23,6,7). These drug-metabolizing enzymes affect the pharmacokinetics of many medications. Some medications are converted to an active form by these enzymes, and some are inactivated. The contributions of CYP enzymes to the pharmacokinetics of neuropsychiatric medications have been well-described; however, there is less evidence on whether variants in these genes are associated with treatment efficacy, especially in pediatric patients.8,9 CYP2D6 enzyme activity reaches adult levels soon after birth, but children may have higher CYP2C19 activity than adults.4 CYP3A4 also contributes to the metabolism of many medications; however, there is only weak evidence that genetic variants in CYP3A4 contribute to variability in the pharmacokinetics of these medications, and there are currently no dosing guidelines based on pharmacogenetics available for this gene.10
As is common in the pharmacogenetic field, genotypes are denoted with a “star allele” (eg, *2) rather than positional nomenclature (eg, c.681G>A). The normal allele is usually designated as *1, and this result is given in the absence of the tested alleles. There is no consensus on the minimum set of alleles to be tested for most genes,11 so commercially available tests vary widely in what alleles are tested (and therefore what they exclude before calling a normal allele).12 The metabolizer phenotype for a patient is determined by taking into account the activity of each of the patient’s 2 alleles (eg, *1/*2). A patient is categorized as a poor-, intermediate-, normal- (extensive-), or ultra-rapid metabolizer. Generally, the allele definitions are widely agreed upon (what genetic variant or variants comprise the *2 allele) due to nomenclature committees for each gene; however, because there are no standards for interpretation, the interpretation of the activity of the alleles and conversion to metabolizer phenotype varies among clinics.13
Continue to: Guidelines help with genotype-guided dosing