User login
Leveraging the Million Veteran Program Infrastructure and Data for a Rapid Research Response to COVID-19
The Million Veteran Program (MVP) was launched in 2011 by the US Department of Veterans Affairs (VA) to enroll at least 1 million veterans in a longitudinal cohort to better understand how genes, lifestyle, military experience, and environmental exposures interact to influence health and illness and ultimately enable precision health care. The MVP has established a national, centralized infrastructure for recruitment and enrollment, biospecimen and data collection and storage, data generation and curation, and secure data access. When the COVID-19 pandemic hit in 2020, the MVP was leveraged to support research utilizing the following key infrastructure components: (1) MVP recruitment and enrollment platform to provide support for COVID-19 vaccine and treatment trials and to collect COVID-19 data from MVP participants; (2) using MVP Phenomics for COVID-19 research data cleaning and curation, assisting with the development of a VA Severity Index for COVID-19, and forming 6 scientific working groups to coordinate COVID-19 research questions; and (3) the VA/MVP and US Department of Energy (DOE) partnership to assist in responding to COVID-19 research questions identified by the US Food and Drug Administration (FDA). This article describes these infrastructure components in more detail and highlights key findings from the MVP COVID-19 research efforts.
MVP Infrastructure
The Veterans Health Administration (VHA) Office of Research and Development (ORD) oversaw efforts to develop the VA Coronavirus Research Volunteer List (the COVID-19 registry). To support the registry, the MVP leveraged its infrastructure to facilitate a rapid response. The MVP is designed as a full-service and centralized recruitment and enrollment platform. This includes MVP office oversight; MVP coordinating centers that manage the centralized platform; an information center that handles inbound and outbound calls; an informatics system built for recruitment and enrollment monitoring and tracking; and a network of more than 70 participating MVP sites with dedicated staff to conduct recruitment and enrollment activities. The MVP used its informatics infrastructure to support secure data storage for the registry volunteer information. MVP coordinating center staff worked with the COVID-19 registry to invite > 125,000 MVP participants from approximately 20 MVP sites. Additionally, MVP information center staff made > 4000 calls to prospective registry volunteers. This work resulted in 1300 volunteers agreeing to be
New Data Collection
The MVP protocol was approved by the VA Central Institutional Review Board (IRB) in 2011. As part of initial enrollment in MVP, participants consented to recontact for additional self-report information along with access to their electronic health record (EHR). This allows for the linkage of EHR and survey response data, thus providing a comprehensive understanding of health history before and after a self-reported COVID-19 diagnosis. Between May 2020 and September 2021, the MVP COVID-19 survey was distributed to existing MVP participants via mail, telephone, and email with the ability to complete the survey by paper and pencil or through the MVP online system. Dissemination of the survey was approved by the VA Central IRB in 2020, with nearly 730,000 eligible MVP participants contacted. As of June 2022, 255,737 MVP participants (35% of the eligible cohort) had completed the survey; 86% completed a paper survey while 14% completed it online. Respondents were primarily older (≥ 65 years); 90% were male; close to 7% reported Hispanic ethnicity, and 11% reported Black race.
Findings from this survey provide insight into pandemic behaviors not consistently captured in EHRs, such as psychosocial aspects, including social and emotional support, loss of tangible and intangible resources, as well as COVID-19–related behaviors, such as social distancing and self-protective practices.1 MVP COVID-19 survey data combined with veteran EHRs, responses to other MVP surveys, and genetic data enable MVP researchers to better understand epidemiological, clinical, and psychosocial aspects of the disease. Future COVID-19 studies may use self-reported survey responses to enrich understanding about the effects of the disease on a veteran’s daily life, and possibly validate existing EHR COVID-19 diagnoses and hospitalization findings. This comprehensive data resource provides a unique opportunity to identify new targets for disease prevention, treatment, and management with an emphasis on individual variability in genes, environment, and lifestyle.
COVID-19 Research
In early 2020, the burden of COVID-19 on the US was unprecedented, and little was known about risk factors for severe COVID-19 and deaths. The MVP Phenomics team quickly responded with a large-scale phenome-wide association study (PheWAS) of >
To broaden disease progression data curation and fit the specific needs of the VA, we operationalized and validated the World Health Organization clinical severity scale and used VA EHR data to create the VA Severity Index for COVID-19 (VASIC).3 The VASIC category is now part of the MVP core data repository, where volumes of data from multiple activities are integrated through an automated process to create monthly research-ready data cubes. These activities include extensive data curation, mapping, phenotyping, and adjudication that are performed to curate oxygen supplementation status and other procedures related to treatment that are processed and understood in real time. The data cubes were provisioned to MVP COVID-19 researchers. In addition, the VASIC scale variable is now integrated within the larger VA system for all researchers to use as part of its wider COVID-19 initiative. The VA Centralized Interactive Phenomics Resource (CIPHER) phenomics library now hosts the details of VASIC, codes, metadata, and related COVID-19 data products for all VA communities. In partnership with CIPHER and other internal and external COVID-19 initiatives, the MVP continues to play an integral part for the VA and beyond in the development of a phenomics algorithm for long COVID, or post-acute COVID-19 syndrome (PACS).
Host Genetics in COVID-19
As the SARS-CoV-2 virus continued to spread globally, it became clear that the symptoms and severity of infection experienced by patients varied across a broad spectrum, from being asymptomatic carriers to experiencing severe symptoms in 1 or more organ systems in the body, resulting in death. This variability suggested that host genetics and other host factors may play a role in determining the severity of COVID-19 infection. The MVP dataset, with genetic and health information on > 600,000 MVP participants, provided an ideal dataset to explore host contributions to COVID-19.
In late spring 2020, the MVP executive committee issued a call to the MVP research community to propose study aims around the COVID-19 pandemic that could leverage the phenotypic and genetic data and resources. The MVP quickly formed 6 rapid-response scientific working groups. Their mission was to cultivate collaboration and inclusivity and to coordinate COVID-19 research questions. A steering committee composed of the MVP executive committee, staff from computational environments, working group cochairs, and an administrator, who was responsible for daily oversight of the working groups. In addition, the ORD COVID-19 steering committee reviewed and approved research activities to ensure scientific rigor, as well as alignment with overall ongoing research activities.
The MVP COVID-19 working groups included dozens of researchers who used MVP data to identify disease mechanisms; understand the impact of host genetics on susceptibility, morbidity, and mortality; and identify potential targets for treatments and therapies. The working groups were further supported by MVP analysts to work cross-functionally on genomics, phenomics, statistical genetics, and PheWAS. Each working group chair was responsible for prioritizing concepts and moving them forward in coordination with the MVP and ORD COVID-19 steering committees. An overview of the MVP COVID-19 working groups follows (Table).4-9
Druggable genome. This working group researched drug-repurposing opportunities to prevent severe COVID-19, defined as hospitalization with oxygen therapy (high flow), intubation, mechanical ventilation, vasopressors, dialysis, or death from COVID-19; and prevent complications in patients hospitalized by COVID-19.
Pharmacogenomics. This working group focused on 2 main aims: the impact of apolipoprotein L1 risk variants on acute kidney injury (AKI) and death in Black veterans with COVID-19; and pharmacogenetic analysis of remdesivir-induced liver chemistry abnormalities.
Disease mechanisms. Understanding the underlying pathways and mechanisms behind COVID-19 has been a difficult but important challenge overall in the scientific community. This working group investigated specific genetic markers and effects on COVID-19, including polygenic predisposition to venous thromboembolism associated with increased COVID-19 susceptibility; renal comorbidities and new AKI and unfavorable outcomes among COVID-19–positive sickle cell trait carriers; and mucin 5B, oligomeric mucus/gel-forming gene polymorphism, and protective effects in COVID-19 infection.
Genomics for risk prediction, polygenic risk scores, and mendelian randomization. Risk prediction for COVID-19 has been widely studied mostly aiming at comorbidities and preexisting conditions. The MVP cohort provided a unique opportunity to understand how genetic information can enhance our understanding of COVID-19 risk. This working group focused on: (1) ABO blood group typing and the protective effects of the O blood group on COVID-19 infection; (2) polygenic risk scores and COVID-19 outcomes; (3) human leukocyte antigen typing and COVID-19 outcomes; and (4) a transcriptome-wide association study of COVID-19–positive MVP participants.
Genome-Wide Association Study (GWAS) and Downstream Analysis. This working group performed GWAS of the main COVID-19 outcomes. Results from GWAS unveiled new genetic loci to suggest further investigation on these candidate genes. The results were used by other MVP COVID-19 working groups for their activities. The results also contributed to external collaborations, such as the COVID-19 Host Genetics Initiative.
COVID-19–Related PheWAS. This working group focused on understanding the potential clinical significance of genetic variants associated with susceptibility to, or outcomes of, COVID-19 infection. They worked to identify traits that share genetic variants associated with severe COVID-19 from the Host Genetics Initiative. The group also studied the phenotypic consequences of acquired mosaic chromosomal alterations with early data linking to COVID-19 susceptibility.
COVID-19 Research Partnerships
In 2016, the VA and DOE formed an interagency partnership known as Computational Health Analytics for Medical Precision to Improve Outcomes Now (CHAMPION) to demonstrate the power of combining the VA EHR system, MVP genetic data, and clinical research expertise with DOE high-performance computing infrastructure and artificial intelligence expertise. The VA EHR captures longitudinal care information on veterans with records that go back decades. Furthermore, the VA covers the costs of medications and
The DOE Oak Ridge National Laboratory (ORNL) in Tennessee securely maintains this rich database for the VA. The ORNL Summit supercomputer can complete trillions of calculations per second to provide critical and timely analyses, applying the most advanced and powerful artificial intelligence methods, which would not be possible in more conventional research settings. CHAMPION taught the VA and DOE how to bring their disparate research cultures together for innovative collaborative investigation. Moreover, this collaboration produced a cadre of VA and DOE scientists familiar with VA patient data and experienced in conducting joint research successfully and integrating omics data with clinical data for a better mechanistic understanding. Because of this preexisting collaboration between the VA and DOE, interagency teams were prepared at the start of the COVID-19 pandemic.10-15
Other recently completed studies have developed and validated short-term mortality indices in individuals with COVID-19 based on their preexisting conditions, assessed the generalizability of VA COVID-19 experiences to the US population, and evaluated the effectiveness of hydroxychloroquine with and without azithromycin in VA patients with COVID-19.12,15 A recent study demonstrated the benefit of prophylactic anticoagulation at initial hospitalization.14
The VA also provided the FDA with daily reports on aggregate VA COVID-19 cases and their distribution across the VA system, demographics of VA patients with COVID-19, and analyses of predictive models for positive test results and death. The VA regularly sent the FDA aggregated data showing patterns of medication use and retrospective analyses of the effectiveness of certain medications (including remdesivir and some antithrombotic agents). The FDA used these data along with other data to understand the scope of the pandemic and to predict drug shortages or needs for additional medical equipment, including ventilators.
Limitations
For the most part, MVP infrastructure and partnerships were efficiently leveraged to significantly advance our understanding of the biological basis of COVID-19 and to develop treatments and vaccines. However, there were a few limitations that may have slowed timely and optimal outcomes. An issue not limited to the MVP or VA was the continual evolution of the pandemic and its response. This included evolving definitions of disease, symptomatology, testing, vaccines, and public health recommendations. Keeping pace with the emerging knowledge from these domains was a struggle for the entire scientific community. A more discrete limitation was the number of participants in the MVP with positive COVID-19 test results and positive symptoms; however, this was mitigated by partnering with other groups like the COVID-19 Host Genetics Initiative to increase study participant numbers. Finally, there were logistical and regulatory challenges associated with coordination of national clinical trial recruitment across a VA system with > 100 discrete hospitals.
Conclusions
Having a centralized infrastructure for recruitment and enrollment, including a national research volunteer registry, information center, research staff, and coordinating centers, can allow for expedited enrollment in vaccine and treatment trials in the face of future public health emergencies. VA assets, including its rich EHR and MVP, the world’s largest genomic cohort, have contributed to improving our understanding and management of COVID-19.
1. Whitbourne SB, Nguyen XT, Song RJ, et al. Million Veteran Program’s response to COVID-19: survey development and preliminary findings. PLoS One. 2022;17(4):e0266381. doi:10.1371/journal.pone.0266381
2. Song RJ, Ho YL, Schubert P, et al. Phenome-wide association of 1809 phenotypes and COVID-19 disease progression in the Veterans Health Administration Million Veteran Program. PLoS One. 2021;16(5):e0251651. doi:10.1371/journal.pone.0251651
3. Galloway A, Park Y, Tanukonda V, et al. Impact of COVID-19 severity on long-term events in US veterans using the Veterans Affairs Severity Index for COVID-19 (VASIC). J Infect Dis. 2022;226(12):2113-2117. doi:10.1093/infdis/jiac182
4. Gaziano L, Giambartolomei C, Pereira AC, et al. Actionable druggable genome-wide Mendelian randomization identifies repurposing opportunities for COVID-19. Nat Med. 2021;27(4):668-676. doi:10.0138/s41591-021-01310-z
5. Hung AM, Sha SC, Bick AG, et al. APOL1 risk variants, acute kidney injury, and death in participants with African ancestry hospitalized with COVID-19 from the Million Veteran Program. JAMA Intern Med. 2022;182(4):386-395. doi:10.1001/jamainternmed.2021.8538
6. Verma A, Huffman JE, Gao L, et al. Association of kidney comorbidities and acute kidney failure with unfavorable outcomes after COVID-19 in individuals with the sickle cell trait. JAMA Intern Med. 2022;182(8):796-804. doi:10.1001/jamainternmed.2022.2141
7. Verma A, Tsao NL, Thomann LO, et al. A phenome-wide association study of genes associated with COVID-19 severity reveals shared genetics with complex diseases in the Million Veteran Program. PLoS Genet. 2022;18(4):e1010113. doi:10.1371/journal.pgen.1010113
8. Peloso GM, Tcheandjieu C, McGeary JE, et al. Genetic loci associated with COVID-19 positivity and hospitalization in White, Black, and Hispanic Veterans of the VA Million Veteran Program. Front Genetic. 2022;12:777076. doi:10.3389/fgene.2021.777076
9. Verma A, Minnier J, Wan ES, et al. A MUC5B gene polymorphism, rs35705950-T confers protective effects against COVID-19 hospitalization but not severe disease or mortality. Am J Respir Crit Care Med. 2022;182(8):796-804. doi:10.1164/rccm.202109-2166OC
10. Garvin MR, Alvarez C, Miller JI, et al. A mechanistic model and therapeutic interventions for COVID-19 involving a RAS-mediated bradykinin storm. Elife. 2020;e59177. doi:10.7554/eLife.59177
11. Rentsch CT, Kidwai-Khan F, Tate JP, et al. Patterns of COVID-19 testing and mortality by race and ethnicity among United States veterans: A nationwide cohort study. PLoS Med. 2020;17(9):e1003379. doi:10.1371/journal.pmed.1003379
12. King JT, Yoon JS, Rentsch CT, et al. Development and validation of a 30-day mortality index based on pre-existing medical administrative data from 13,323 COVID-19 patients: the Veterans Health Administration COVID-19 (VACO) Index. PLoS One. 2020;15(11):e0241825. doi:10.1371/journal.pone.0241825
13. Joubert W, Weighill D, Kainer D, et al. Attacking the opioid epidemic: determining the epistatic and pleiotropic genetic architectures for chronic pain and opioid addiction. SC18: International Conference for High Performance Computing, Networking, Storage and Analysis. Dallas, TX, USA, 2018:717-730. doi:10.1109/SC.2018.00060
14. Rentsch CT, Beckman JA, Tomlinson L, et al. Early initiation of prophylactic anticoagulation for prevention of COVID-19 mortality: a nationwide cohort study of hospitalized patients in the United States. BMJ. 2021;372:n311. doi:10.1136/bmj.n311
15. Gerlovin H, Posner DC, Ho YL, et al. Pharmacoepidemiology, machine learning, and COVID-19: an intent-to-treat analysis of hydroxychloroquine, with or without Azithromycin, and COVID-19 outcomes among hospitalized US Veterans. Am J Epidemiol. 2021;190(11): 2405-2419. doi:10.1093/aje/kwab183
The Million Veteran Program (MVP) was launched in 2011 by the US Department of Veterans Affairs (VA) to enroll at least 1 million veterans in a longitudinal cohort to better understand how genes, lifestyle, military experience, and environmental exposures interact to influence health and illness and ultimately enable precision health care. The MVP has established a national, centralized infrastructure for recruitment and enrollment, biospecimen and data collection and storage, data generation and curation, and secure data access. When the COVID-19 pandemic hit in 2020, the MVP was leveraged to support research utilizing the following key infrastructure components: (1) MVP recruitment and enrollment platform to provide support for COVID-19 vaccine and treatment trials and to collect COVID-19 data from MVP participants; (2) using MVP Phenomics for COVID-19 research data cleaning and curation, assisting with the development of a VA Severity Index for COVID-19, and forming 6 scientific working groups to coordinate COVID-19 research questions; and (3) the VA/MVP and US Department of Energy (DOE) partnership to assist in responding to COVID-19 research questions identified by the US Food and Drug Administration (FDA). This article describes these infrastructure components in more detail and highlights key findings from the MVP COVID-19 research efforts.
MVP Infrastructure
The Veterans Health Administration (VHA) Office of Research and Development (ORD) oversaw efforts to develop the VA Coronavirus Research Volunteer List (the COVID-19 registry). To support the registry, the MVP leveraged its infrastructure to facilitate a rapid response. The MVP is designed as a full-service and centralized recruitment and enrollment platform. This includes MVP office oversight; MVP coordinating centers that manage the centralized platform; an information center that handles inbound and outbound calls; an informatics system built for recruitment and enrollment monitoring and tracking; and a network of more than 70 participating MVP sites with dedicated staff to conduct recruitment and enrollment activities. The MVP used its informatics infrastructure to support secure data storage for the registry volunteer information. MVP coordinating center staff worked with the COVID-19 registry to invite > 125,000 MVP participants from approximately 20 MVP sites. Additionally, MVP information center staff made > 4000 calls to prospective registry volunteers. This work resulted in 1300 volunteers agreeing to be
New Data Collection
The MVP protocol was approved by the VA Central Institutional Review Board (IRB) in 2011. As part of initial enrollment in MVP, participants consented to recontact for additional self-report information along with access to their electronic health record (EHR). This allows for the linkage of EHR and survey response data, thus providing a comprehensive understanding of health history before and after a self-reported COVID-19 diagnosis. Between May 2020 and September 2021, the MVP COVID-19 survey was distributed to existing MVP participants via mail, telephone, and email with the ability to complete the survey by paper and pencil or through the MVP online system. Dissemination of the survey was approved by the VA Central IRB in 2020, with nearly 730,000 eligible MVP participants contacted. As of June 2022, 255,737 MVP participants (35% of the eligible cohort) had completed the survey; 86% completed a paper survey while 14% completed it online. Respondents were primarily older (≥ 65 years); 90% were male; close to 7% reported Hispanic ethnicity, and 11% reported Black race.
Findings from this survey provide insight into pandemic behaviors not consistently captured in EHRs, such as psychosocial aspects, including social and emotional support, loss of tangible and intangible resources, as well as COVID-19–related behaviors, such as social distancing and self-protective practices.1 MVP COVID-19 survey data combined with veteran EHRs, responses to other MVP surveys, and genetic data enable MVP researchers to better understand epidemiological, clinical, and psychosocial aspects of the disease. Future COVID-19 studies may use self-reported survey responses to enrich understanding about the effects of the disease on a veteran’s daily life, and possibly validate existing EHR COVID-19 diagnoses and hospitalization findings. This comprehensive data resource provides a unique opportunity to identify new targets for disease prevention, treatment, and management with an emphasis on individual variability in genes, environment, and lifestyle.
COVID-19 Research
In early 2020, the burden of COVID-19 on the US was unprecedented, and little was known about risk factors for severe COVID-19 and deaths. The MVP Phenomics team quickly responded with a large-scale phenome-wide association study (PheWAS) of >
To broaden disease progression data curation and fit the specific needs of the VA, we operationalized and validated the World Health Organization clinical severity scale and used VA EHR data to create the VA Severity Index for COVID-19 (VASIC).3 The VASIC category is now part of the MVP core data repository, where volumes of data from multiple activities are integrated through an automated process to create monthly research-ready data cubes. These activities include extensive data curation, mapping, phenotyping, and adjudication that are performed to curate oxygen supplementation status and other procedures related to treatment that are processed and understood in real time. The data cubes were provisioned to MVP COVID-19 researchers. In addition, the VASIC scale variable is now integrated within the larger VA system for all researchers to use as part of its wider COVID-19 initiative. The VA Centralized Interactive Phenomics Resource (CIPHER) phenomics library now hosts the details of VASIC, codes, metadata, and related COVID-19 data products for all VA communities. In partnership with CIPHER and other internal and external COVID-19 initiatives, the MVP continues to play an integral part for the VA and beyond in the development of a phenomics algorithm for long COVID, or post-acute COVID-19 syndrome (PACS).
Host Genetics in COVID-19
As the SARS-CoV-2 virus continued to spread globally, it became clear that the symptoms and severity of infection experienced by patients varied across a broad spectrum, from being asymptomatic carriers to experiencing severe symptoms in 1 or more organ systems in the body, resulting in death. This variability suggested that host genetics and other host factors may play a role in determining the severity of COVID-19 infection. The MVP dataset, with genetic and health information on > 600,000 MVP participants, provided an ideal dataset to explore host contributions to COVID-19.
In late spring 2020, the MVP executive committee issued a call to the MVP research community to propose study aims around the COVID-19 pandemic that could leverage the phenotypic and genetic data and resources. The MVP quickly formed 6 rapid-response scientific working groups. Their mission was to cultivate collaboration and inclusivity and to coordinate COVID-19 research questions. A steering committee composed of the MVP executive committee, staff from computational environments, working group cochairs, and an administrator, who was responsible for daily oversight of the working groups. In addition, the ORD COVID-19 steering committee reviewed and approved research activities to ensure scientific rigor, as well as alignment with overall ongoing research activities.
The MVP COVID-19 working groups included dozens of researchers who used MVP data to identify disease mechanisms; understand the impact of host genetics on susceptibility, morbidity, and mortality; and identify potential targets for treatments and therapies. The working groups were further supported by MVP analysts to work cross-functionally on genomics, phenomics, statistical genetics, and PheWAS. Each working group chair was responsible for prioritizing concepts and moving them forward in coordination with the MVP and ORD COVID-19 steering committees. An overview of the MVP COVID-19 working groups follows (Table).4-9
Druggable genome. This working group researched drug-repurposing opportunities to prevent severe COVID-19, defined as hospitalization with oxygen therapy (high flow), intubation, mechanical ventilation, vasopressors, dialysis, or death from COVID-19; and prevent complications in patients hospitalized by COVID-19.
Pharmacogenomics. This working group focused on 2 main aims: the impact of apolipoprotein L1 risk variants on acute kidney injury (AKI) and death in Black veterans with COVID-19; and pharmacogenetic analysis of remdesivir-induced liver chemistry abnormalities.
Disease mechanisms. Understanding the underlying pathways and mechanisms behind COVID-19 has been a difficult but important challenge overall in the scientific community. This working group investigated specific genetic markers and effects on COVID-19, including polygenic predisposition to venous thromboembolism associated with increased COVID-19 susceptibility; renal comorbidities and new AKI and unfavorable outcomes among COVID-19–positive sickle cell trait carriers; and mucin 5B, oligomeric mucus/gel-forming gene polymorphism, and protective effects in COVID-19 infection.
Genomics for risk prediction, polygenic risk scores, and mendelian randomization. Risk prediction for COVID-19 has been widely studied mostly aiming at comorbidities and preexisting conditions. The MVP cohort provided a unique opportunity to understand how genetic information can enhance our understanding of COVID-19 risk. This working group focused on: (1) ABO blood group typing and the protective effects of the O blood group on COVID-19 infection; (2) polygenic risk scores and COVID-19 outcomes; (3) human leukocyte antigen typing and COVID-19 outcomes; and (4) a transcriptome-wide association study of COVID-19–positive MVP participants.
Genome-Wide Association Study (GWAS) and Downstream Analysis. This working group performed GWAS of the main COVID-19 outcomes. Results from GWAS unveiled new genetic loci to suggest further investigation on these candidate genes. The results were used by other MVP COVID-19 working groups for their activities. The results also contributed to external collaborations, such as the COVID-19 Host Genetics Initiative.
COVID-19–Related PheWAS. This working group focused on understanding the potential clinical significance of genetic variants associated with susceptibility to, or outcomes of, COVID-19 infection. They worked to identify traits that share genetic variants associated with severe COVID-19 from the Host Genetics Initiative. The group also studied the phenotypic consequences of acquired mosaic chromosomal alterations with early data linking to COVID-19 susceptibility.
COVID-19 Research Partnerships
In 2016, the VA and DOE formed an interagency partnership known as Computational Health Analytics for Medical Precision to Improve Outcomes Now (CHAMPION) to demonstrate the power of combining the VA EHR system, MVP genetic data, and clinical research expertise with DOE high-performance computing infrastructure and artificial intelligence expertise. The VA EHR captures longitudinal care information on veterans with records that go back decades. Furthermore, the VA covers the costs of medications and
The DOE Oak Ridge National Laboratory (ORNL) in Tennessee securely maintains this rich database for the VA. The ORNL Summit supercomputer can complete trillions of calculations per second to provide critical and timely analyses, applying the most advanced and powerful artificial intelligence methods, which would not be possible in more conventional research settings. CHAMPION taught the VA and DOE how to bring their disparate research cultures together for innovative collaborative investigation. Moreover, this collaboration produced a cadre of VA and DOE scientists familiar with VA patient data and experienced in conducting joint research successfully and integrating omics data with clinical data for a better mechanistic understanding. Because of this preexisting collaboration between the VA and DOE, interagency teams were prepared at the start of the COVID-19 pandemic.10-15
Other recently completed studies have developed and validated short-term mortality indices in individuals with COVID-19 based on their preexisting conditions, assessed the generalizability of VA COVID-19 experiences to the US population, and evaluated the effectiveness of hydroxychloroquine with and without azithromycin in VA patients with COVID-19.12,15 A recent study demonstrated the benefit of prophylactic anticoagulation at initial hospitalization.14
The VA also provided the FDA with daily reports on aggregate VA COVID-19 cases and their distribution across the VA system, demographics of VA patients with COVID-19, and analyses of predictive models for positive test results and death. The VA regularly sent the FDA aggregated data showing patterns of medication use and retrospective analyses of the effectiveness of certain medications (including remdesivir and some antithrombotic agents). The FDA used these data along with other data to understand the scope of the pandemic and to predict drug shortages or needs for additional medical equipment, including ventilators.
Limitations
For the most part, MVP infrastructure and partnerships were efficiently leveraged to significantly advance our understanding of the biological basis of COVID-19 and to develop treatments and vaccines. However, there were a few limitations that may have slowed timely and optimal outcomes. An issue not limited to the MVP or VA was the continual evolution of the pandemic and its response. This included evolving definitions of disease, symptomatology, testing, vaccines, and public health recommendations. Keeping pace with the emerging knowledge from these domains was a struggle for the entire scientific community. A more discrete limitation was the number of participants in the MVP with positive COVID-19 test results and positive symptoms; however, this was mitigated by partnering with other groups like the COVID-19 Host Genetics Initiative to increase study participant numbers. Finally, there were logistical and regulatory challenges associated with coordination of national clinical trial recruitment across a VA system with > 100 discrete hospitals.
Conclusions
Having a centralized infrastructure for recruitment and enrollment, including a national research volunteer registry, information center, research staff, and coordinating centers, can allow for expedited enrollment in vaccine and treatment trials in the face of future public health emergencies. VA assets, including its rich EHR and MVP, the world’s largest genomic cohort, have contributed to improving our understanding and management of COVID-19.
The Million Veteran Program (MVP) was launched in 2011 by the US Department of Veterans Affairs (VA) to enroll at least 1 million veterans in a longitudinal cohort to better understand how genes, lifestyle, military experience, and environmental exposures interact to influence health and illness and ultimately enable precision health care. The MVP has established a national, centralized infrastructure for recruitment and enrollment, biospecimen and data collection and storage, data generation and curation, and secure data access. When the COVID-19 pandemic hit in 2020, the MVP was leveraged to support research utilizing the following key infrastructure components: (1) MVP recruitment and enrollment platform to provide support for COVID-19 vaccine and treatment trials and to collect COVID-19 data from MVP participants; (2) using MVP Phenomics for COVID-19 research data cleaning and curation, assisting with the development of a VA Severity Index for COVID-19, and forming 6 scientific working groups to coordinate COVID-19 research questions; and (3) the VA/MVP and US Department of Energy (DOE) partnership to assist in responding to COVID-19 research questions identified by the US Food and Drug Administration (FDA). This article describes these infrastructure components in more detail and highlights key findings from the MVP COVID-19 research efforts.
MVP Infrastructure
The Veterans Health Administration (VHA) Office of Research and Development (ORD) oversaw efforts to develop the VA Coronavirus Research Volunteer List (the COVID-19 registry). To support the registry, the MVP leveraged its infrastructure to facilitate a rapid response. The MVP is designed as a full-service and centralized recruitment and enrollment platform. This includes MVP office oversight; MVP coordinating centers that manage the centralized platform; an information center that handles inbound and outbound calls; an informatics system built for recruitment and enrollment monitoring and tracking; and a network of more than 70 participating MVP sites with dedicated staff to conduct recruitment and enrollment activities. The MVP used its informatics infrastructure to support secure data storage for the registry volunteer information. MVP coordinating center staff worked with the COVID-19 registry to invite > 125,000 MVP participants from approximately 20 MVP sites. Additionally, MVP information center staff made > 4000 calls to prospective registry volunteers. This work resulted in 1300 volunteers agreeing to be
New Data Collection
The MVP protocol was approved by the VA Central Institutional Review Board (IRB) in 2011. As part of initial enrollment in MVP, participants consented to recontact for additional self-report information along with access to their electronic health record (EHR). This allows for the linkage of EHR and survey response data, thus providing a comprehensive understanding of health history before and after a self-reported COVID-19 diagnosis. Between May 2020 and September 2021, the MVP COVID-19 survey was distributed to existing MVP participants via mail, telephone, and email with the ability to complete the survey by paper and pencil or through the MVP online system. Dissemination of the survey was approved by the VA Central IRB in 2020, with nearly 730,000 eligible MVP participants contacted. As of June 2022, 255,737 MVP participants (35% of the eligible cohort) had completed the survey; 86% completed a paper survey while 14% completed it online. Respondents were primarily older (≥ 65 years); 90% were male; close to 7% reported Hispanic ethnicity, and 11% reported Black race.
Findings from this survey provide insight into pandemic behaviors not consistently captured in EHRs, such as psychosocial aspects, including social and emotional support, loss of tangible and intangible resources, as well as COVID-19–related behaviors, such as social distancing and self-protective practices.1 MVP COVID-19 survey data combined with veteran EHRs, responses to other MVP surveys, and genetic data enable MVP researchers to better understand epidemiological, clinical, and psychosocial aspects of the disease. Future COVID-19 studies may use self-reported survey responses to enrich understanding about the effects of the disease on a veteran’s daily life, and possibly validate existing EHR COVID-19 diagnoses and hospitalization findings. This comprehensive data resource provides a unique opportunity to identify new targets for disease prevention, treatment, and management with an emphasis on individual variability in genes, environment, and lifestyle.
COVID-19 Research
In early 2020, the burden of COVID-19 on the US was unprecedented, and little was known about risk factors for severe COVID-19 and deaths. The MVP Phenomics team quickly responded with a large-scale phenome-wide association study (PheWAS) of >
To broaden disease progression data curation and fit the specific needs of the VA, we operationalized and validated the World Health Organization clinical severity scale and used VA EHR data to create the VA Severity Index for COVID-19 (VASIC).3 The VASIC category is now part of the MVP core data repository, where volumes of data from multiple activities are integrated through an automated process to create monthly research-ready data cubes. These activities include extensive data curation, mapping, phenotyping, and adjudication that are performed to curate oxygen supplementation status and other procedures related to treatment that are processed and understood in real time. The data cubes were provisioned to MVP COVID-19 researchers. In addition, the VASIC scale variable is now integrated within the larger VA system for all researchers to use as part of its wider COVID-19 initiative. The VA Centralized Interactive Phenomics Resource (CIPHER) phenomics library now hosts the details of VASIC, codes, metadata, and related COVID-19 data products for all VA communities. In partnership with CIPHER and other internal and external COVID-19 initiatives, the MVP continues to play an integral part for the VA and beyond in the development of a phenomics algorithm for long COVID, or post-acute COVID-19 syndrome (PACS).
Host Genetics in COVID-19
As the SARS-CoV-2 virus continued to spread globally, it became clear that the symptoms and severity of infection experienced by patients varied across a broad spectrum, from being asymptomatic carriers to experiencing severe symptoms in 1 or more organ systems in the body, resulting in death. This variability suggested that host genetics and other host factors may play a role in determining the severity of COVID-19 infection. The MVP dataset, with genetic and health information on > 600,000 MVP participants, provided an ideal dataset to explore host contributions to COVID-19.
In late spring 2020, the MVP executive committee issued a call to the MVP research community to propose study aims around the COVID-19 pandemic that could leverage the phenotypic and genetic data and resources. The MVP quickly formed 6 rapid-response scientific working groups. Their mission was to cultivate collaboration and inclusivity and to coordinate COVID-19 research questions. A steering committee composed of the MVP executive committee, staff from computational environments, working group cochairs, and an administrator, who was responsible for daily oversight of the working groups. In addition, the ORD COVID-19 steering committee reviewed and approved research activities to ensure scientific rigor, as well as alignment with overall ongoing research activities.
The MVP COVID-19 working groups included dozens of researchers who used MVP data to identify disease mechanisms; understand the impact of host genetics on susceptibility, morbidity, and mortality; and identify potential targets for treatments and therapies. The working groups were further supported by MVP analysts to work cross-functionally on genomics, phenomics, statistical genetics, and PheWAS. Each working group chair was responsible for prioritizing concepts and moving them forward in coordination with the MVP and ORD COVID-19 steering committees. An overview of the MVP COVID-19 working groups follows (Table).4-9
Druggable genome. This working group researched drug-repurposing opportunities to prevent severe COVID-19, defined as hospitalization with oxygen therapy (high flow), intubation, mechanical ventilation, vasopressors, dialysis, or death from COVID-19; and prevent complications in patients hospitalized by COVID-19.
Pharmacogenomics. This working group focused on 2 main aims: the impact of apolipoprotein L1 risk variants on acute kidney injury (AKI) and death in Black veterans with COVID-19; and pharmacogenetic analysis of remdesivir-induced liver chemistry abnormalities.
Disease mechanisms. Understanding the underlying pathways and mechanisms behind COVID-19 has been a difficult but important challenge overall in the scientific community. This working group investigated specific genetic markers and effects on COVID-19, including polygenic predisposition to venous thromboembolism associated with increased COVID-19 susceptibility; renal comorbidities and new AKI and unfavorable outcomes among COVID-19–positive sickle cell trait carriers; and mucin 5B, oligomeric mucus/gel-forming gene polymorphism, and protective effects in COVID-19 infection.
Genomics for risk prediction, polygenic risk scores, and mendelian randomization. Risk prediction for COVID-19 has been widely studied mostly aiming at comorbidities and preexisting conditions. The MVP cohort provided a unique opportunity to understand how genetic information can enhance our understanding of COVID-19 risk. This working group focused on: (1) ABO blood group typing and the protective effects of the O blood group on COVID-19 infection; (2) polygenic risk scores and COVID-19 outcomes; (3) human leukocyte antigen typing and COVID-19 outcomes; and (4) a transcriptome-wide association study of COVID-19–positive MVP participants.
Genome-Wide Association Study (GWAS) and Downstream Analysis. This working group performed GWAS of the main COVID-19 outcomes. Results from GWAS unveiled new genetic loci to suggest further investigation on these candidate genes. The results were used by other MVP COVID-19 working groups for their activities. The results also contributed to external collaborations, such as the COVID-19 Host Genetics Initiative.
COVID-19–Related PheWAS. This working group focused on understanding the potential clinical significance of genetic variants associated with susceptibility to, or outcomes of, COVID-19 infection. They worked to identify traits that share genetic variants associated with severe COVID-19 from the Host Genetics Initiative. The group also studied the phenotypic consequences of acquired mosaic chromosomal alterations with early data linking to COVID-19 susceptibility.
COVID-19 Research Partnerships
In 2016, the VA and DOE formed an interagency partnership known as Computational Health Analytics for Medical Precision to Improve Outcomes Now (CHAMPION) to demonstrate the power of combining the VA EHR system, MVP genetic data, and clinical research expertise with DOE high-performance computing infrastructure and artificial intelligence expertise. The VA EHR captures longitudinal care information on veterans with records that go back decades. Furthermore, the VA covers the costs of medications and
The DOE Oak Ridge National Laboratory (ORNL) in Tennessee securely maintains this rich database for the VA. The ORNL Summit supercomputer can complete trillions of calculations per second to provide critical and timely analyses, applying the most advanced and powerful artificial intelligence methods, which would not be possible in more conventional research settings. CHAMPION taught the VA and DOE how to bring their disparate research cultures together for innovative collaborative investigation. Moreover, this collaboration produced a cadre of VA and DOE scientists familiar with VA patient data and experienced in conducting joint research successfully and integrating omics data with clinical data for a better mechanistic understanding. Because of this preexisting collaboration between the VA and DOE, interagency teams were prepared at the start of the COVID-19 pandemic.10-15
Other recently completed studies have developed and validated short-term mortality indices in individuals with COVID-19 based on their preexisting conditions, assessed the generalizability of VA COVID-19 experiences to the US population, and evaluated the effectiveness of hydroxychloroquine with and without azithromycin in VA patients with COVID-19.12,15 A recent study demonstrated the benefit of prophylactic anticoagulation at initial hospitalization.14
The VA also provided the FDA with daily reports on aggregate VA COVID-19 cases and their distribution across the VA system, demographics of VA patients with COVID-19, and analyses of predictive models for positive test results and death. The VA regularly sent the FDA aggregated data showing patterns of medication use and retrospective analyses of the effectiveness of certain medications (including remdesivir and some antithrombotic agents). The FDA used these data along with other data to understand the scope of the pandemic and to predict drug shortages or needs for additional medical equipment, including ventilators.
Limitations
For the most part, MVP infrastructure and partnerships were efficiently leveraged to significantly advance our understanding of the biological basis of COVID-19 and to develop treatments and vaccines. However, there were a few limitations that may have slowed timely and optimal outcomes. An issue not limited to the MVP or VA was the continual evolution of the pandemic and its response. This included evolving definitions of disease, symptomatology, testing, vaccines, and public health recommendations. Keeping pace with the emerging knowledge from these domains was a struggle for the entire scientific community. A more discrete limitation was the number of participants in the MVP with positive COVID-19 test results and positive symptoms; however, this was mitigated by partnering with other groups like the COVID-19 Host Genetics Initiative to increase study participant numbers. Finally, there were logistical and regulatory challenges associated with coordination of national clinical trial recruitment across a VA system with > 100 discrete hospitals.
Conclusions
Having a centralized infrastructure for recruitment and enrollment, including a national research volunteer registry, information center, research staff, and coordinating centers, can allow for expedited enrollment in vaccine and treatment trials in the face of future public health emergencies. VA assets, including its rich EHR and MVP, the world’s largest genomic cohort, have contributed to improving our understanding and management of COVID-19.
1. Whitbourne SB, Nguyen XT, Song RJ, et al. Million Veteran Program’s response to COVID-19: survey development and preliminary findings. PLoS One. 2022;17(4):e0266381. doi:10.1371/journal.pone.0266381
2. Song RJ, Ho YL, Schubert P, et al. Phenome-wide association of 1809 phenotypes and COVID-19 disease progression in the Veterans Health Administration Million Veteran Program. PLoS One. 2021;16(5):e0251651. doi:10.1371/journal.pone.0251651
3. Galloway A, Park Y, Tanukonda V, et al. Impact of COVID-19 severity on long-term events in US veterans using the Veterans Affairs Severity Index for COVID-19 (VASIC). J Infect Dis. 2022;226(12):2113-2117. doi:10.1093/infdis/jiac182
4. Gaziano L, Giambartolomei C, Pereira AC, et al. Actionable druggable genome-wide Mendelian randomization identifies repurposing opportunities for COVID-19. Nat Med. 2021;27(4):668-676. doi:10.0138/s41591-021-01310-z
5. Hung AM, Sha SC, Bick AG, et al. APOL1 risk variants, acute kidney injury, and death in participants with African ancestry hospitalized with COVID-19 from the Million Veteran Program. JAMA Intern Med. 2022;182(4):386-395. doi:10.1001/jamainternmed.2021.8538
6. Verma A, Huffman JE, Gao L, et al. Association of kidney comorbidities and acute kidney failure with unfavorable outcomes after COVID-19 in individuals with the sickle cell trait. JAMA Intern Med. 2022;182(8):796-804. doi:10.1001/jamainternmed.2022.2141
7. Verma A, Tsao NL, Thomann LO, et al. A phenome-wide association study of genes associated with COVID-19 severity reveals shared genetics with complex diseases in the Million Veteran Program. PLoS Genet. 2022;18(4):e1010113. doi:10.1371/journal.pgen.1010113
8. Peloso GM, Tcheandjieu C, McGeary JE, et al. Genetic loci associated with COVID-19 positivity and hospitalization in White, Black, and Hispanic Veterans of the VA Million Veteran Program. Front Genetic. 2022;12:777076. doi:10.3389/fgene.2021.777076
9. Verma A, Minnier J, Wan ES, et al. A MUC5B gene polymorphism, rs35705950-T confers protective effects against COVID-19 hospitalization but not severe disease or mortality. Am J Respir Crit Care Med. 2022;182(8):796-804. doi:10.1164/rccm.202109-2166OC
10. Garvin MR, Alvarez C, Miller JI, et al. A mechanistic model and therapeutic interventions for COVID-19 involving a RAS-mediated bradykinin storm. Elife. 2020;e59177. doi:10.7554/eLife.59177
11. Rentsch CT, Kidwai-Khan F, Tate JP, et al. Patterns of COVID-19 testing and mortality by race and ethnicity among United States veterans: A nationwide cohort study. PLoS Med. 2020;17(9):e1003379. doi:10.1371/journal.pmed.1003379
12. King JT, Yoon JS, Rentsch CT, et al. Development and validation of a 30-day mortality index based on pre-existing medical administrative data from 13,323 COVID-19 patients: the Veterans Health Administration COVID-19 (VACO) Index. PLoS One. 2020;15(11):e0241825. doi:10.1371/journal.pone.0241825
13. Joubert W, Weighill D, Kainer D, et al. Attacking the opioid epidemic: determining the epistatic and pleiotropic genetic architectures for chronic pain and opioid addiction. SC18: International Conference for High Performance Computing, Networking, Storage and Analysis. Dallas, TX, USA, 2018:717-730. doi:10.1109/SC.2018.00060
14. Rentsch CT, Beckman JA, Tomlinson L, et al. Early initiation of prophylactic anticoagulation for prevention of COVID-19 mortality: a nationwide cohort study of hospitalized patients in the United States. BMJ. 2021;372:n311. doi:10.1136/bmj.n311
15. Gerlovin H, Posner DC, Ho YL, et al. Pharmacoepidemiology, machine learning, and COVID-19: an intent-to-treat analysis of hydroxychloroquine, with or without Azithromycin, and COVID-19 outcomes among hospitalized US Veterans. Am J Epidemiol. 2021;190(11): 2405-2419. doi:10.1093/aje/kwab183
1. Whitbourne SB, Nguyen XT, Song RJ, et al. Million Veteran Program’s response to COVID-19: survey development and preliminary findings. PLoS One. 2022;17(4):e0266381. doi:10.1371/journal.pone.0266381
2. Song RJ, Ho YL, Schubert P, et al. Phenome-wide association of 1809 phenotypes and COVID-19 disease progression in the Veterans Health Administration Million Veteran Program. PLoS One. 2021;16(5):e0251651. doi:10.1371/journal.pone.0251651
3. Galloway A, Park Y, Tanukonda V, et al. Impact of COVID-19 severity on long-term events in US veterans using the Veterans Affairs Severity Index for COVID-19 (VASIC). J Infect Dis. 2022;226(12):2113-2117. doi:10.1093/infdis/jiac182
4. Gaziano L, Giambartolomei C, Pereira AC, et al. Actionable druggable genome-wide Mendelian randomization identifies repurposing opportunities for COVID-19. Nat Med. 2021;27(4):668-676. doi:10.0138/s41591-021-01310-z
5. Hung AM, Sha SC, Bick AG, et al. APOL1 risk variants, acute kidney injury, and death in participants with African ancestry hospitalized with COVID-19 from the Million Veteran Program. JAMA Intern Med. 2022;182(4):386-395. doi:10.1001/jamainternmed.2021.8538
6. Verma A, Huffman JE, Gao L, et al. Association of kidney comorbidities and acute kidney failure with unfavorable outcomes after COVID-19 in individuals with the sickle cell trait. JAMA Intern Med. 2022;182(8):796-804. doi:10.1001/jamainternmed.2022.2141
7. Verma A, Tsao NL, Thomann LO, et al. A phenome-wide association study of genes associated with COVID-19 severity reveals shared genetics with complex diseases in the Million Veteran Program. PLoS Genet. 2022;18(4):e1010113. doi:10.1371/journal.pgen.1010113
8. Peloso GM, Tcheandjieu C, McGeary JE, et al. Genetic loci associated with COVID-19 positivity and hospitalization in White, Black, and Hispanic Veterans of the VA Million Veteran Program. Front Genetic. 2022;12:777076. doi:10.3389/fgene.2021.777076
9. Verma A, Minnier J, Wan ES, et al. A MUC5B gene polymorphism, rs35705950-T confers protective effects against COVID-19 hospitalization but not severe disease or mortality. Am J Respir Crit Care Med. 2022;182(8):796-804. doi:10.1164/rccm.202109-2166OC
10. Garvin MR, Alvarez C, Miller JI, et al. A mechanistic model and therapeutic interventions for COVID-19 involving a RAS-mediated bradykinin storm. Elife. 2020;e59177. doi:10.7554/eLife.59177
11. Rentsch CT, Kidwai-Khan F, Tate JP, et al. Patterns of COVID-19 testing and mortality by race and ethnicity among United States veterans: A nationwide cohort study. PLoS Med. 2020;17(9):e1003379. doi:10.1371/journal.pmed.1003379
12. King JT, Yoon JS, Rentsch CT, et al. Development and validation of a 30-day mortality index based on pre-existing medical administrative data from 13,323 COVID-19 patients: the Veterans Health Administration COVID-19 (VACO) Index. PLoS One. 2020;15(11):e0241825. doi:10.1371/journal.pone.0241825
13. Joubert W, Weighill D, Kainer D, et al. Attacking the opioid epidemic: determining the epistatic and pleiotropic genetic architectures for chronic pain and opioid addiction. SC18: International Conference for High Performance Computing, Networking, Storage and Analysis. Dallas, TX, USA, 2018:717-730. doi:10.1109/SC.2018.00060
14. Rentsch CT, Beckman JA, Tomlinson L, et al. Early initiation of prophylactic anticoagulation for prevention of COVID-19 mortality: a nationwide cohort study of hospitalized patients in the United States. BMJ. 2021;372:n311. doi:10.1136/bmj.n311
15. Gerlovin H, Posner DC, Ho YL, et al. Pharmacoepidemiology, machine learning, and COVID-19: an intent-to-treat analysis of hydroxychloroquine, with or without Azithromycin, and COVID-19 outcomes among hospitalized US Veterans. Am J Epidemiol. 2021;190(11): 2405-2419. doi:10.1093/aje/kwab183
The Precision Oncology Program for Cancer of the Prostate (POPCaP) Network: A Veterans Affairs/Prostate Cancer Foundation Collaboration(FULL)
The US Department of Veterans Affairs (VA) is home to the Veterans Health Administration (VHA), which delivers care at 1,255 health care facilities, including 170 medical centers. The VA serves 6 million veterans each year and is the largest integrated provider of cancer care in the US. The system uses a single, enterprise-wide electronic health record. The detailed curation of clinical outcomes, laboratory results, and radiology is used in VA efforts to improve oncology outcomes for veterans. The VA also has a National Precision Oncology Program (NPOP), which offers system-wide DNA sequencing for veterans with cancer. Given its size, integration, and capabilities, the VA is an ideal setting for rapid learning cycles of testing and implementing best practices at scale.
Prostate cancer is the most common malignancy affecting men in the US. It is the most commonly-diagnosed solid tumor in the VA, and in 2014, there were 11,376 prostate cancer diagnoses in the VA.1 The clinical characteristics and treatment of veterans with prostate cancer largely parallel the broader population of men in the US.1 Although the majority of men diagnosed with prostate cancer have disease localized to the prostate, an important minority develop metastatic disease, which represents a risk for substantial morbidity and is the lethal form of the disease. Research has yielded transformative advances in the care of men with metastatic prostate cancer, including drugs targeting the testosterone/androgen signaling axis, taxane chemotherapy, the radionuclide radium-223, and a dendritic cell vaccine. Unfortunately, the magnitude and duration of response to these therapies varies widely, and determining the biology relevant to an individual patient that would better inform their treatment decisions is a critical next step. As the ability to interrogate the cancer genome has improved, relevant drivers of tumorigenesis and predictive biomarkers are being identified rapidly, and oncology care has evolved from a one-size-fits-all approach to a precision approach, which uses these biomarkers to assist in therapeutic decision making.
Precision Oncology for Prostate Cancer
A series of studies interrogating the genomics of metastatic prostate cancer have been critical to defining the relevance of precision oncology for prostate cancer. Most of what is known about the genomics of prostate cancer has been derived from analysis of samples from the prostate itself. These samples may not reflect the biology of metastasis and genetic evolution in response to treatment pressure, so the genomic alterations in metastatic disease remained incompletely characterized. Two large research teams supported by grants from the American Association for Cancer Research, Stand Up 2 Cancer, and Prostate Cancer Foundation (PCF) focused their efforts on sampling and analyzing metastatic tissue to define the most relevant genomic alterations in advanced prostate cancer.
These efforts defined a broad range of relatively common alterations in the androgen receptor, as well as the tumor suppressors TP53 and PTEN.2,3 Important subsets of less common alterations in pathways that were potentially targetable were also found, including new alterations in PIK3CA/B, BRAF/RAF1, and β-catenin. Most surprisingly, alterations of DNA repair pathways, including mismatch repair and homologous recombination were found in 20% of tumors, and half of these tumors contained germline alterations. The same groups performed a follow up analysis of germline DNA from men with metastatic prostate cancer, which confirmed that 12% of these patients carry a pathogenic germline alteration in a DNA repair pathway gene.4 These efforts immediately invigorated precision oncology clinical trials for prostate cancer and spurred an effort to find the molecular alterations that could be leveraged to improve care for men with advanced prostate cancer.
Targetable Alterations
Currently a number of genomic alterations are immediately actionable. There are several agents approved by the US Food and Drug Administration (FDA) that exploit these Achilles heels of prostate cancer. Mismatch repair deficiency occurs when any of a group of genes responsible for proofreading the fidelity of DNA replication is compromised by mutation or deletion. Imperfect reading and correction subsequently lead to many DNA mutations in a tissue (hypermutation), which then increases the risk of developing malignancy. If a defective gene in the mismatch repair pathway is inherited, a patient has a genetic predisposition to specific malignancies that are part of the Lynch syndrome.5 Prostate cancer is a relatively rare manifestation of Lynch syndrome, although it is considered one of the malignancies in the Lynch syndrome spectrum.6
Alteration of one of the mismatch repair genes also can occur spontaneously in a tumor, resulting in the same high frequency of spontaneous DNA mutations. Overall, between 3% and 5% of metastatic prostate cancers contain mismatch repair deficiency. The majority of these cases are a result of spontaneous loss or mutation of the relevant gene, but 1 in 5 of these tumors occurs as a component of Lynch syndrome.7 Identification of mismatch repair deficiency is critical because the resulting hypermutation makes these tumors particularly susceptible to intervention with immunotherapy. Up to half of patients with metastatic prostate cancer can have durable responses. This finding is consistent with the experience treating other malignancies with mismatch repair deficiency.8 Although screening for mismatch repair deficiency is standard of care for patients with malignancies such as colorectal cancer, few patients with prostate cancer may receive the mismatch repair deficiency screening (based on unpublished data). In contrast, screening is routine for patients with adenocarcinoma of the lung because their proportion of ROS1 and ALK alterations is similar to the frequency of mismatch repair deficiency when compared with patients with prostate cancer.9
Homologous recombination is another mechanism by which cells repair DNA damage and is responsible for repairing double strand breaks, the type of DNA damage most likely to lead to carcinogenesis. In advanced prostate cancer, BRCA2, ATM, BRCA1 and other members of the Fanconi Anemia/BRCA gene family are altered 20% of the time. These genes also are the most common germline alterations implicated in the development of prostate cancer.2,10 Prostate cancer is considered a BRCA-related cancer much like breast, ovarian, and pancreatic cancers. Defects in homologous recombination repair make BRCA-altered prostate cancers susceptible to DNA damaging chemotherapy, such as platinum and to the use of poly–(adenosine diphosphate–ribose) polymerase (PARP) inhibitors because cancer cells then accumulate cytotoxic and apoptotic levels of DNA.11
In May 2020, the FDA approved the use of PARP inhibitors for the treatment of prostate cancers that contain BRCA and other DNA repair alterations. Rucaparib received accelerated approval for the treatment of prostate cancers containing BRCA alterations and olaparib received full approval for treatment of prostate cancers containing an array of alterations in DNA repair genes.12,13 Both approvals were the direct result of the cited landmark studies that demonstrated the frequency of these alterations in advanced prostate cancer.2,3
Beyond mismatch and homologous recombination repair, there are a large number of potentially targetable alterations found in advanced prostate cancer. It is thus critical that we put systems into place both to find germline and somatic alterations that will inform a veteran’s clinical care and to provide veterans access to precision oncology clinical trials.
The POPCaP Network
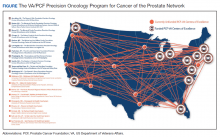
Because prostate cancer is such a significant issue in the VA and best practices for precision oncology can be implemented broadly once defined as successful, the PCF and the VA formed a collaboration to support a network of centers that would focus on implementing a comprehensive strategy for precision oncology in prostate cancer. There are currently 11 centers in the Precision Oncology Program for Cancer of the Prostate (POPCaP) network (Figure). These centers are tasked with comprehensively sequencing germline and somatic tissue from veterans with metastatic prostate cancer to find alterations, which could provide access to treatments that would otherwise not be available or appropriate.
The network is collaborating with NPOP, which provides clinical grade tumor gene panel sequencing for veterans with prostate cancer from > 90% of VA medical centers. POPCaP also partners with the University of Washington to use its OncoPlex gene panel and University of Michigan to use the Oncomine panel to define the best platform for defining targetable alterations for veterans with prostate cancer. Investigators participate in a monthly molecular oncology tumor board continuing medical education-accredited program, which provides guidance and education across the VA about the evidence available to assist in decision making for veterans sequenced through NPOP and the academic platforms. These efforts leverage VA’s partnership with IBM Watson for Genomics to annotate DNA sequencing results to provide clinicians with potential therapeutic options for veterans.
A clinical trials mechanism is embedded in POPCaP to broaden treatment options, improve care for men with prostate cancer, and leverage the sequencing efforts in the network. The Prostate Cancer Analysis for Therapy Choice (PATCH) clinical trials network employs an umbrella study approach whereby alterations are identified through sequencing and veterans are given access to studies embedded at sites across the network. Graff and Huang provide a detailed description of the PATCH network and its potential as a multisite clinical trials mechanism.14 For studies within the network, funds can be provided to support travel to participate in clinical trials for veterans who would be eligible for study but do not live in a catchment for a network site. POPCaP also leverages both the resources of the National Cancer Institute (NCI)-designated cancer centers that are VA academic affiliates, as well as a VA/NCI partnership (NAVIGATE) to increase veteran access to NCI cutting-edge clinical trials.
The network has regular teleconference meetings of the investigators, coordinators, and stakeholders and face-to-face meetings, which are coordinated around other national meetings. These meetings enable investigators to work collaboratively to advance current knowledge in prostate cancer through the application of complementary and synergistic research approaches. Since research plays a critical role within the learning health care system, POPCaP investigators are working to optimize the transfer of knowledge from the clinic to the bench and back to the clinic. In this regard, investigators from network sites have organized themselves into working groups to focus on multiple critical aspects of research and care within the network, including sequencing, phenotyping, health services, health disparities, and a network biorepository.
VA Office of Research and Development
With support from the VA Office of Research and Development, there are research efforts focused on the development of data analytics to identify veterans with metastatic prostate cancer within the electronic health record to ensure access to appropriate testing, treatment, and clinical trials. This will optimize tracking and continuous quality improvement in precision oncology. The Office of Research and Development also supports the use of artificial intelligence to identify predictive markers for diagnosis, prognosis, therapeutic response and patient stratification. POPCaP investigators, along with other investigators from across the VA, conduct research that continually improves the care of veterans with prostate cancer. POPCaP has a special focus on prostate cancer among African Americans, who are disproportionately affected by the disease and well represented in VA. The efforts of the working groups, the research studies and the network as a whole also serve to recruit both junior and senior investigators to the VA in order to support the VA research enterprise.
Active collaborations between the network and other elements of VA include efforts to optimize germline testing and genetic counseling in prostate cancer through the Genomic Medicine Service, which provides telehealth genetic counseling throughout the VA. POPCaP pilots innovative approaches to increase access to clinical genetics and genetic counseling services to support the volume of genetic testing of veterans with cancer. Current National Comprehensive Cancer Network (NCCN) guidelines recommend germline testing for all men with metastatic prostate cancer, which can efficiently identify the roughly 10% of veterans with metastatic disease who carry a germline alteration and provide them with access to studies, FDA-approved treatments, while also offering critical health care information to family members who may also carry a pathogenic germline alteration.
Million Veteran Program
The Million Veteran Program (MVP) has collected > 825,000 germline DNA samples from an anticipated enrollment of > 1 million veterans in one of the most ambitious genetic research efforts to correlate how germline DNA interacts with lifestyle, medications and military exposure to affect health and illness (www.research.va.gov/mvp). MVP is a racially and ethnically diverse veteran cohort that is roughly 20% African American and 7% Hispanic. More than 40,000 of the participants have had prostate cancer, one third of whom are African Americans, giving researchers unprecedented ability to discover factors that impact the development and treatment of the disease in this population. In particular, MVP will provide unique insights into the genetic mutations that drive the development of aggressive prostate cancer in all male veterans, including African Americans. These discoveries will undoubtedly lead to improved screening of and treatment for prostate cancer.
In order to demonstrate clinical utility as well as the infrastructure needs to scale up within the VHA, MVP has launched a pilot project that offers to return clinically actionable genetic results to MVP participants with metastatic prostate cancer, opening the door to new therapies to improve the length and quality of these veterans’ lives. Importantly, the pilot includes cascade testing in family members of enrolled veterans. Given that the original MVP consent did not allow for return of results, and MVP genetic testing is research grade, veterans who volunteer will provide a second consent and undergo clinical genetic testing to confirm the variants. Results from this pilot study also will inform expansion of VA precision oncology efforts for patients with other cancers such as breast cancer or ovarian cancer, where the specific genetic mutations are known to play a role, (eg, BRCA2). In addition, through an interagency agreement with the US Department of Energy (DOE), MVP is leveraging DOE expertise and high-performance computing capabilities to identify clinical and genetic risk factors for prostate cancer that will progress to metastatic disease.
This active research collaboration between POPCaP, MVP, and the Genomic Medicine Service will identify germline BRCA alterations from MVP participants with metastatic prostate cancer and give them access to therapies that may provide better outcomes and access to genetic testing for their family members.
Future Directions
The POPCaP network and its partnership with VA clinical and research efforts is anticipated to provide important insights into barriers and solutions to the implementation of precision oncology for prostate cancer across the VA. These lessons learned may also be relevant for precision oncology care in other settings. As an example, the role of germline testing and genetic counseling is growing more relevant in precision oncology, yet it is clear that the number of men and women dealing with malignancy who actually receive counseling and testing is suboptimal in most health care systems.14 Optimizing the quality and efficiency of oncogenetics within the VA system in a manner that gives access to these services for every veteran in urban or rural environments is an important goal.
The VA has done extensive work in teleoncology and the Genomic Medicine Service provides telehealth genetic counseling service to 90 VA medical facilities nationwide. Expanding on this model to create a distributed network system across the country is an opportunity that will continue to raise VA profile as a leader in this area while providing increased access to genetic services.
Finally, the clinical trials network within POPCaP already has provided valuable insights into how research efforts that originate within the VA can leverage the VA’s strengths. The use of the NPOP centralized sequencing platform to identify potentially targetable alterations across medical centers provides the potential to bring critical access to research to veterans where they live through virtual clinical trials. The VA has a centralized institutional review board that can service large multisite study participation efficiently across the VA. The promise of virtual clinical trials to interrogate relatively rare biomarkers would benefit from institution of a virtual clinical trials workflow. In theory patients with a potentially targetable biomarker could be identified through the centralized DNA sequencing platform and a clinical trial team of virtual investigators and research coordinators would work with health care providers at sites for study startup and performance. Efforts to design and implement this approach are actively being pursued.
The goal of the VA/PCF POPCaP network is to make certain that every veteran has access to appropriate genetic and genomic testing and that the results are utilized so that veterans with targetable alterations receive the best clinical care and have access to clinical trials that could benefit them individually while advancing knowledge that benefits all.
1. Montgomery B, Williams C. Prostate cancer federal health care data trends. https://www.mdedge.com/fedprac/article/208077/oncology/prostate-cancer-federal-health-care-data-trends. Published September 1, 2019. Accessed July 16, 2020.
2. Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer [published correction appears in Cell. 2015 Jul 16;162(2):454]. Cell. 2015;161(5):1215-1228. doi:10.1016/j.cell.2015.05.001
3. Quigley DA, Dang HX, Zhao SG, et al. Genomic hallmarks and structural variation in metastatic prostate cancer [published correction appears in Cell. 2018 Oct 18;175(3):889]. Cell. 2018;174(3):758-769.e9. doi:10.1016/j.cell.2018.06.039
4. Pritchard CC, Offit K, Nelson PS. DNA-repair gene mutations in metastatic prostate cancer. N Engl J Med. 2016;375(18):1804-1805. doi:10.1056/NEJMc1611137
5. Guillem JG. Molecular diagnosis of hereditary nonpolyposis colon cancer. N Engl J Med. 1998;339(13):924-925. doi:10.1056/nejm199809243391316
6. Ryan S, Jenkins MA, Win AK. Risk of prostate cancer in Lynch syndrome: a systematic review and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2014;23(3):437-449. doi:10.1158/1055-9965.EPI-13-1165
7. Abida W, Cheng ML, Armenia J, et al. Analysis of the prevalence of microsatellite instability in prostate cancer and response to immune checkpoint blockade. JAMA Oncol. 2019;5(4):471-478. doi:10.1001/jamaoncol.2018.5801
8. Graham LS, Montgomery B, Cheng HH, et al. Mismatch repair deficiency in metastatic prostate cancer: Response to PD-1 blockade and standard therapies. PLoS One. 2020;15(5):e0233260. Published 2020 May 26. doi:10.1371/journal.pone.0233260
9. Yu HA, Planchard D, Lovly CM. Sequencing therapy for genetically defined subgroups of non-small cell lung cancer. Am Soc Clin Oncol Educ Book. 2018;38:726-739. doi:10.1200/EDBK_201331
10. Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375(5):443-453. doi:10.1056/NEJMoa1603144
11. Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917-921. doi:10.1038/nature03445
12. Abida W, Campbell D, Patnaik A, et al. Preliminary results from the TRITON2 study of rucaparib in patients with DNA damage repair deficiency metastatic, castration resistant prostate cancer: updated analyses. Ann Oncol. 2019;30(suppl 5): v325-v355. doi:10.1093/annonc/mdz248
13. de Bono J, Mateo J, Fizazi K, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382(22):2091-2102. doi:10.1056/NEJMoa1911440
14. Graff JN, Huang GD. Leveraging Veterans Health Administration clinical and research resources to accelerate discovery and testing in precision oncology. Fed Pract. 2020;37(suppl 4):S62-S67. doi: 10.12788/fp.0028
The US Department of Veterans Affairs (VA) is home to the Veterans Health Administration (VHA), which delivers care at 1,255 health care facilities, including 170 medical centers. The VA serves 6 million veterans each year and is the largest integrated provider of cancer care in the US. The system uses a single, enterprise-wide electronic health record. The detailed curation of clinical outcomes, laboratory results, and radiology is used in VA efforts to improve oncology outcomes for veterans. The VA also has a National Precision Oncology Program (NPOP), which offers system-wide DNA sequencing for veterans with cancer. Given its size, integration, and capabilities, the VA is an ideal setting for rapid learning cycles of testing and implementing best practices at scale.
Prostate cancer is the most common malignancy affecting men in the US. It is the most commonly-diagnosed solid tumor in the VA, and in 2014, there were 11,376 prostate cancer diagnoses in the VA.1 The clinical characteristics and treatment of veterans with prostate cancer largely parallel the broader population of men in the US.1 Although the majority of men diagnosed with prostate cancer have disease localized to the prostate, an important minority develop metastatic disease, which represents a risk for substantial morbidity and is the lethal form of the disease. Research has yielded transformative advances in the care of men with metastatic prostate cancer, including drugs targeting the testosterone/androgen signaling axis, taxane chemotherapy, the radionuclide radium-223, and a dendritic cell vaccine. Unfortunately, the magnitude and duration of response to these therapies varies widely, and determining the biology relevant to an individual patient that would better inform their treatment decisions is a critical next step. As the ability to interrogate the cancer genome has improved, relevant drivers of tumorigenesis and predictive biomarkers are being identified rapidly, and oncology care has evolved from a one-size-fits-all approach to a precision approach, which uses these biomarkers to assist in therapeutic decision making.
Precision Oncology for Prostate Cancer
A series of studies interrogating the genomics of metastatic prostate cancer have been critical to defining the relevance of precision oncology for prostate cancer. Most of what is known about the genomics of prostate cancer has been derived from analysis of samples from the prostate itself. These samples may not reflect the biology of metastasis and genetic evolution in response to treatment pressure, so the genomic alterations in metastatic disease remained incompletely characterized. Two large research teams supported by grants from the American Association for Cancer Research, Stand Up 2 Cancer, and Prostate Cancer Foundation (PCF) focused their efforts on sampling and analyzing metastatic tissue to define the most relevant genomic alterations in advanced prostate cancer.
These efforts defined a broad range of relatively common alterations in the androgen receptor, as well as the tumor suppressors TP53 and PTEN.2,3 Important subsets of less common alterations in pathways that were potentially targetable were also found, including new alterations in PIK3CA/B, BRAF/RAF1, and β-catenin. Most surprisingly, alterations of DNA repair pathways, including mismatch repair and homologous recombination were found in 20% of tumors, and half of these tumors contained germline alterations. The same groups performed a follow up analysis of germline DNA from men with metastatic prostate cancer, which confirmed that 12% of these patients carry a pathogenic germline alteration in a DNA repair pathway gene.4 These efforts immediately invigorated precision oncology clinical trials for prostate cancer and spurred an effort to find the molecular alterations that could be leveraged to improve care for men with advanced prostate cancer.
Targetable Alterations
Currently a number of genomic alterations are immediately actionable. There are several agents approved by the US Food and Drug Administration (FDA) that exploit these Achilles heels of prostate cancer. Mismatch repair deficiency occurs when any of a group of genes responsible for proofreading the fidelity of DNA replication is compromised by mutation or deletion. Imperfect reading and correction subsequently lead to many DNA mutations in a tissue (hypermutation), which then increases the risk of developing malignancy. If a defective gene in the mismatch repair pathway is inherited, a patient has a genetic predisposition to specific malignancies that are part of the Lynch syndrome.5 Prostate cancer is a relatively rare manifestation of Lynch syndrome, although it is considered one of the malignancies in the Lynch syndrome spectrum.6
Alteration of one of the mismatch repair genes also can occur spontaneously in a tumor, resulting in the same high frequency of spontaneous DNA mutations. Overall, between 3% and 5% of metastatic prostate cancers contain mismatch repair deficiency. The majority of these cases are a result of spontaneous loss or mutation of the relevant gene, but 1 in 5 of these tumors occurs as a component of Lynch syndrome.7 Identification of mismatch repair deficiency is critical because the resulting hypermutation makes these tumors particularly susceptible to intervention with immunotherapy. Up to half of patients with metastatic prostate cancer can have durable responses. This finding is consistent with the experience treating other malignancies with mismatch repair deficiency.8 Although screening for mismatch repair deficiency is standard of care for patients with malignancies such as colorectal cancer, few patients with prostate cancer may receive the mismatch repair deficiency screening (based on unpublished data). In contrast, screening is routine for patients with adenocarcinoma of the lung because their proportion of ROS1 and ALK alterations is similar to the frequency of mismatch repair deficiency when compared with patients with prostate cancer.9
Homologous recombination is another mechanism by which cells repair DNA damage and is responsible for repairing double strand breaks, the type of DNA damage most likely to lead to carcinogenesis. In advanced prostate cancer, BRCA2, ATM, BRCA1 and other members of the Fanconi Anemia/BRCA gene family are altered 20% of the time. These genes also are the most common germline alterations implicated in the development of prostate cancer.2,10 Prostate cancer is considered a BRCA-related cancer much like breast, ovarian, and pancreatic cancers. Defects in homologous recombination repair make BRCA-altered prostate cancers susceptible to DNA damaging chemotherapy, such as platinum and to the use of poly–(adenosine diphosphate–ribose) polymerase (PARP) inhibitors because cancer cells then accumulate cytotoxic and apoptotic levels of DNA.11
In May 2020, the FDA approved the use of PARP inhibitors for the treatment of prostate cancers that contain BRCA and other DNA repair alterations. Rucaparib received accelerated approval for the treatment of prostate cancers containing BRCA alterations and olaparib received full approval for treatment of prostate cancers containing an array of alterations in DNA repair genes.12,13 Both approvals were the direct result of the cited landmark studies that demonstrated the frequency of these alterations in advanced prostate cancer.2,3
Beyond mismatch and homologous recombination repair, there are a large number of potentially targetable alterations found in advanced prostate cancer. It is thus critical that we put systems into place both to find germline and somatic alterations that will inform a veteran’s clinical care and to provide veterans access to precision oncology clinical trials.
The POPCaP Network
Because prostate cancer is such a significant issue in the VA and best practices for precision oncology can be implemented broadly once defined as successful, the PCF and the VA formed a collaboration to support a network of centers that would focus on implementing a comprehensive strategy for precision oncology in prostate cancer. There are currently 11 centers in the Precision Oncology Program for Cancer of the Prostate (POPCaP) network (Figure). These centers are tasked with comprehensively sequencing germline and somatic tissue from veterans with metastatic prostate cancer to find alterations, which could provide access to treatments that would otherwise not be available or appropriate.
The network is collaborating with NPOP, which provides clinical grade tumor gene panel sequencing for veterans with prostate cancer from > 90% of VA medical centers. POPCaP also partners with the University of Washington to use its OncoPlex gene panel and University of Michigan to use the Oncomine panel to define the best platform for defining targetable alterations for veterans with prostate cancer. Investigators participate in a monthly molecular oncology tumor board continuing medical education-accredited program, which provides guidance and education across the VA about the evidence available to assist in decision making for veterans sequenced through NPOP and the academic platforms. These efforts leverage VA’s partnership with IBM Watson for Genomics to annotate DNA sequencing results to provide clinicians with potential therapeutic options for veterans.
A clinical trials mechanism is embedded in POPCaP to broaden treatment options, improve care for men with prostate cancer, and leverage the sequencing efforts in the network. The Prostate Cancer Analysis for Therapy Choice (PATCH) clinical trials network employs an umbrella study approach whereby alterations are identified through sequencing and veterans are given access to studies embedded at sites across the network. Graff and Huang provide a detailed description of the PATCH network and its potential as a multisite clinical trials mechanism.14 For studies within the network, funds can be provided to support travel to participate in clinical trials for veterans who would be eligible for study but do not live in a catchment for a network site. POPCaP also leverages both the resources of the National Cancer Institute (NCI)-designated cancer centers that are VA academic affiliates, as well as a VA/NCI partnership (NAVIGATE) to increase veteran access to NCI cutting-edge clinical trials.
The network has regular teleconference meetings of the investigators, coordinators, and stakeholders and face-to-face meetings, which are coordinated around other national meetings. These meetings enable investigators to work collaboratively to advance current knowledge in prostate cancer through the application of complementary and synergistic research approaches. Since research plays a critical role within the learning health care system, POPCaP investigators are working to optimize the transfer of knowledge from the clinic to the bench and back to the clinic. In this regard, investigators from network sites have organized themselves into working groups to focus on multiple critical aspects of research and care within the network, including sequencing, phenotyping, health services, health disparities, and a network biorepository.
VA Office of Research and Development
With support from the VA Office of Research and Development, there are research efforts focused on the development of data analytics to identify veterans with metastatic prostate cancer within the electronic health record to ensure access to appropriate testing, treatment, and clinical trials. This will optimize tracking and continuous quality improvement in precision oncology. The Office of Research and Development also supports the use of artificial intelligence to identify predictive markers for diagnosis, prognosis, therapeutic response and patient stratification. POPCaP investigators, along with other investigators from across the VA, conduct research that continually improves the care of veterans with prostate cancer. POPCaP has a special focus on prostate cancer among African Americans, who are disproportionately affected by the disease and well represented in VA. The efforts of the working groups, the research studies and the network as a whole also serve to recruit both junior and senior investigators to the VA in order to support the VA research enterprise.
Active collaborations between the network and other elements of VA include efforts to optimize germline testing and genetic counseling in prostate cancer through the Genomic Medicine Service, which provides telehealth genetic counseling throughout the VA. POPCaP pilots innovative approaches to increase access to clinical genetics and genetic counseling services to support the volume of genetic testing of veterans with cancer. Current National Comprehensive Cancer Network (NCCN) guidelines recommend germline testing for all men with metastatic prostate cancer, which can efficiently identify the roughly 10% of veterans with metastatic disease who carry a germline alteration and provide them with access to studies, FDA-approved treatments, while also offering critical health care information to family members who may also carry a pathogenic germline alteration.
Million Veteran Program
The Million Veteran Program (MVP) has collected > 825,000 germline DNA samples from an anticipated enrollment of > 1 million veterans in one of the most ambitious genetic research efforts to correlate how germline DNA interacts with lifestyle, medications and military exposure to affect health and illness (www.research.va.gov/mvp). MVP is a racially and ethnically diverse veteran cohort that is roughly 20% African American and 7% Hispanic. More than 40,000 of the participants have had prostate cancer, one third of whom are African Americans, giving researchers unprecedented ability to discover factors that impact the development and treatment of the disease in this population. In particular, MVP will provide unique insights into the genetic mutations that drive the development of aggressive prostate cancer in all male veterans, including African Americans. These discoveries will undoubtedly lead to improved screening of and treatment for prostate cancer.
In order to demonstrate clinical utility as well as the infrastructure needs to scale up within the VHA, MVP has launched a pilot project that offers to return clinically actionable genetic results to MVP participants with metastatic prostate cancer, opening the door to new therapies to improve the length and quality of these veterans’ lives. Importantly, the pilot includes cascade testing in family members of enrolled veterans. Given that the original MVP consent did not allow for return of results, and MVP genetic testing is research grade, veterans who volunteer will provide a second consent and undergo clinical genetic testing to confirm the variants. Results from this pilot study also will inform expansion of VA precision oncology efforts for patients with other cancers such as breast cancer or ovarian cancer, where the specific genetic mutations are known to play a role, (eg, BRCA2). In addition, through an interagency agreement with the US Department of Energy (DOE), MVP is leveraging DOE expertise and high-performance computing capabilities to identify clinical and genetic risk factors for prostate cancer that will progress to metastatic disease.
This active research collaboration between POPCaP, MVP, and the Genomic Medicine Service will identify germline BRCA alterations from MVP participants with metastatic prostate cancer and give them access to therapies that may provide better outcomes and access to genetic testing for their family members.
Future Directions
The POPCaP network and its partnership with VA clinical and research efforts is anticipated to provide important insights into barriers and solutions to the implementation of precision oncology for prostate cancer across the VA. These lessons learned may also be relevant for precision oncology care in other settings. As an example, the role of germline testing and genetic counseling is growing more relevant in precision oncology, yet it is clear that the number of men and women dealing with malignancy who actually receive counseling and testing is suboptimal in most health care systems.14 Optimizing the quality and efficiency of oncogenetics within the VA system in a manner that gives access to these services for every veteran in urban or rural environments is an important goal.
The VA has done extensive work in teleoncology and the Genomic Medicine Service provides telehealth genetic counseling service to 90 VA medical facilities nationwide. Expanding on this model to create a distributed network system across the country is an opportunity that will continue to raise VA profile as a leader in this area while providing increased access to genetic services.
Finally, the clinical trials network within POPCaP already has provided valuable insights into how research efforts that originate within the VA can leverage the VA’s strengths. The use of the NPOP centralized sequencing platform to identify potentially targetable alterations across medical centers provides the potential to bring critical access to research to veterans where they live through virtual clinical trials. The VA has a centralized institutional review board that can service large multisite study participation efficiently across the VA. The promise of virtual clinical trials to interrogate relatively rare biomarkers would benefit from institution of a virtual clinical trials workflow. In theory patients with a potentially targetable biomarker could be identified through the centralized DNA sequencing platform and a clinical trial team of virtual investigators and research coordinators would work with health care providers at sites for study startup and performance. Efforts to design and implement this approach are actively being pursued.
The goal of the VA/PCF POPCaP network is to make certain that every veteran has access to appropriate genetic and genomic testing and that the results are utilized so that veterans with targetable alterations receive the best clinical care and have access to clinical trials that could benefit them individually while advancing knowledge that benefits all.
The US Department of Veterans Affairs (VA) is home to the Veterans Health Administration (VHA), which delivers care at 1,255 health care facilities, including 170 medical centers. The VA serves 6 million veterans each year and is the largest integrated provider of cancer care in the US. The system uses a single, enterprise-wide electronic health record. The detailed curation of clinical outcomes, laboratory results, and radiology is used in VA efforts to improve oncology outcomes for veterans. The VA also has a National Precision Oncology Program (NPOP), which offers system-wide DNA sequencing for veterans with cancer. Given its size, integration, and capabilities, the VA is an ideal setting for rapid learning cycles of testing and implementing best practices at scale.
Prostate cancer is the most common malignancy affecting men in the US. It is the most commonly-diagnosed solid tumor in the VA, and in 2014, there were 11,376 prostate cancer diagnoses in the VA.1 The clinical characteristics and treatment of veterans with prostate cancer largely parallel the broader population of men in the US.1 Although the majority of men diagnosed with prostate cancer have disease localized to the prostate, an important minority develop metastatic disease, which represents a risk for substantial morbidity and is the lethal form of the disease. Research has yielded transformative advances in the care of men with metastatic prostate cancer, including drugs targeting the testosterone/androgen signaling axis, taxane chemotherapy, the radionuclide radium-223, and a dendritic cell vaccine. Unfortunately, the magnitude and duration of response to these therapies varies widely, and determining the biology relevant to an individual patient that would better inform their treatment decisions is a critical next step. As the ability to interrogate the cancer genome has improved, relevant drivers of tumorigenesis and predictive biomarkers are being identified rapidly, and oncology care has evolved from a one-size-fits-all approach to a precision approach, which uses these biomarkers to assist in therapeutic decision making.
Precision Oncology for Prostate Cancer
A series of studies interrogating the genomics of metastatic prostate cancer have been critical to defining the relevance of precision oncology for prostate cancer. Most of what is known about the genomics of prostate cancer has been derived from analysis of samples from the prostate itself. These samples may not reflect the biology of metastasis and genetic evolution in response to treatment pressure, so the genomic alterations in metastatic disease remained incompletely characterized. Two large research teams supported by grants from the American Association for Cancer Research, Stand Up 2 Cancer, and Prostate Cancer Foundation (PCF) focused their efforts on sampling and analyzing metastatic tissue to define the most relevant genomic alterations in advanced prostate cancer.
These efforts defined a broad range of relatively common alterations in the androgen receptor, as well as the tumor suppressors TP53 and PTEN.2,3 Important subsets of less common alterations in pathways that were potentially targetable were also found, including new alterations in PIK3CA/B, BRAF/RAF1, and β-catenin. Most surprisingly, alterations of DNA repair pathways, including mismatch repair and homologous recombination were found in 20% of tumors, and half of these tumors contained germline alterations. The same groups performed a follow up analysis of germline DNA from men with metastatic prostate cancer, which confirmed that 12% of these patients carry a pathogenic germline alteration in a DNA repair pathway gene.4 These efforts immediately invigorated precision oncology clinical trials for prostate cancer and spurred an effort to find the molecular alterations that could be leveraged to improve care for men with advanced prostate cancer.
Targetable Alterations
Currently a number of genomic alterations are immediately actionable. There are several agents approved by the US Food and Drug Administration (FDA) that exploit these Achilles heels of prostate cancer. Mismatch repair deficiency occurs when any of a group of genes responsible for proofreading the fidelity of DNA replication is compromised by mutation or deletion. Imperfect reading and correction subsequently lead to many DNA mutations in a tissue (hypermutation), which then increases the risk of developing malignancy. If a defective gene in the mismatch repair pathway is inherited, a patient has a genetic predisposition to specific malignancies that are part of the Lynch syndrome.5 Prostate cancer is a relatively rare manifestation of Lynch syndrome, although it is considered one of the malignancies in the Lynch syndrome spectrum.6
Alteration of one of the mismatch repair genes also can occur spontaneously in a tumor, resulting in the same high frequency of spontaneous DNA mutations. Overall, between 3% and 5% of metastatic prostate cancers contain mismatch repair deficiency. The majority of these cases are a result of spontaneous loss or mutation of the relevant gene, but 1 in 5 of these tumors occurs as a component of Lynch syndrome.7 Identification of mismatch repair deficiency is critical because the resulting hypermutation makes these tumors particularly susceptible to intervention with immunotherapy. Up to half of patients with metastatic prostate cancer can have durable responses. This finding is consistent with the experience treating other malignancies with mismatch repair deficiency.8 Although screening for mismatch repair deficiency is standard of care for patients with malignancies such as colorectal cancer, few patients with prostate cancer may receive the mismatch repair deficiency screening (based on unpublished data). In contrast, screening is routine for patients with adenocarcinoma of the lung because their proportion of ROS1 and ALK alterations is similar to the frequency of mismatch repair deficiency when compared with patients with prostate cancer.9
Homologous recombination is another mechanism by which cells repair DNA damage and is responsible for repairing double strand breaks, the type of DNA damage most likely to lead to carcinogenesis. In advanced prostate cancer, BRCA2, ATM, BRCA1 and other members of the Fanconi Anemia/BRCA gene family are altered 20% of the time. These genes also are the most common germline alterations implicated in the development of prostate cancer.2,10 Prostate cancer is considered a BRCA-related cancer much like breast, ovarian, and pancreatic cancers. Defects in homologous recombination repair make BRCA-altered prostate cancers susceptible to DNA damaging chemotherapy, such as platinum and to the use of poly–(adenosine diphosphate–ribose) polymerase (PARP) inhibitors because cancer cells then accumulate cytotoxic and apoptotic levels of DNA.11
In May 2020, the FDA approved the use of PARP inhibitors for the treatment of prostate cancers that contain BRCA and other DNA repair alterations. Rucaparib received accelerated approval for the treatment of prostate cancers containing BRCA alterations and olaparib received full approval for treatment of prostate cancers containing an array of alterations in DNA repair genes.12,13 Both approvals were the direct result of the cited landmark studies that demonstrated the frequency of these alterations in advanced prostate cancer.2,3
Beyond mismatch and homologous recombination repair, there are a large number of potentially targetable alterations found in advanced prostate cancer. It is thus critical that we put systems into place both to find germline and somatic alterations that will inform a veteran’s clinical care and to provide veterans access to precision oncology clinical trials.
The POPCaP Network
Because prostate cancer is such a significant issue in the VA and best practices for precision oncology can be implemented broadly once defined as successful, the PCF and the VA formed a collaboration to support a network of centers that would focus on implementing a comprehensive strategy for precision oncology in prostate cancer. There are currently 11 centers in the Precision Oncology Program for Cancer of the Prostate (POPCaP) network (Figure). These centers are tasked with comprehensively sequencing germline and somatic tissue from veterans with metastatic prostate cancer to find alterations, which could provide access to treatments that would otherwise not be available or appropriate.
The network is collaborating with NPOP, which provides clinical grade tumor gene panel sequencing for veterans with prostate cancer from > 90% of VA medical centers. POPCaP also partners with the University of Washington to use its OncoPlex gene panel and University of Michigan to use the Oncomine panel to define the best platform for defining targetable alterations for veterans with prostate cancer. Investigators participate in a monthly molecular oncology tumor board continuing medical education-accredited program, which provides guidance and education across the VA about the evidence available to assist in decision making for veterans sequenced through NPOP and the academic platforms. These efforts leverage VA’s partnership with IBM Watson for Genomics to annotate DNA sequencing results to provide clinicians with potential therapeutic options for veterans.
A clinical trials mechanism is embedded in POPCaP to broaden treatment options, improve care for men with prostate cancer, and leverage the sequencing efforts in the network. The Prostate Cancer Analysis for Therapy Choice (PATCH) clinical trials network employs an umbrella study approach whereby alterations are identified through sequencing and veterans are given access to studies embedded at sites across the network. Graff and Huang provide a detailed description of the PATCH network and its potential as a multisite clinical trials mechanism.14 For studies within the network, funds can be provided to support travel to participate in clinical trials for veterans who would be eligible for study but do not live in a catchment for a network site. POPCaP also leverages both the resources of the National Cancer Institute (NCI)-designated cancer centers that are VA academic affiliates, as well as a VA/NCI partnership (NAVIGATE) to increase veteran access to NCI cutting-edge clinical trials.
The network has regular teleconference meetings of the investigators, coordinators, and stakeholders and face-to-face meetings, which are coordinated around other national meetings. These meetings enable investigators to work collaboratively to advance current knowledge in prostate cancer through the application of complementary and synergistic research approaches. Since research plays a critical role within the learning health care system, POPCaP investigators are working to optimize the transfer of knowledge from the clinic to the bench and back to the clinic. In this regard, investigators from network sites have organized themselves into working groups to focus on multiple critical aspects of research and care within the network, including sequencing, phenotyping, health services, health disparities, and a network biorepository.
VA Office of Research and Development
With support from the VA Office of Research and Development, there are research efforts focused on the development of data analytics to identify veterans with metastatic prostate cancer within the electronic health record to ensure access to appropriate testing, treatment, and clinical trials. This will optimize tracking and continuous quality improvement in precision oncology. The Office of Research and Development also supports the use of artificial intelligence to identify predictive markers for diagnosis, prognosis, therapeutic response and patient stratification. POPCaP investigators, along with other investigators from across the VA, conduct research that continually improves the care of veterans with prostate cancer. POPCaP has a special focus on prostate cancer among African Americans, who are disproportionately affected by the disease and well represented in VA. The efforts of the working groups, the research studies and the network as a whole also serve to recruit both junior and senior investigators to the VA in order to support the VA research enterprise.
Active collaborations between the network and other elements of VA include efforts to optimize germline testing and genetic counseling in prostate cancer through the Genomic Medicine Service, which provides telehealth genetic counseling throughout the VA. POPCaP pilots innovative approaches to increase access to clinical genetics and genetic counseling services to support the volume of genetic testing of veterans with cancer. Current National Comprehensive Cancer Network (NCCN) guidelines recommend germline testing for all men with metastatic prostate cancer, which can efficiently identify the roughly 10% of veterans with metastatic disease who carry a germline alteration and provide them with access to studies, FDA-approved treatments, while also offering critical health care information to family members who may also carry a pathogenic germline alteration.
Million Veteran Program
The Million Veteran Program (MVP) has collected > 825,000 germline DNA samples from an anticipated enrollment of > 1 million veterans in one of the most ambitious genetic research efforts to correlate how germline DNA interacts with lifestyle, medications and military exposure to affect health and illness (www.research.va.gov/mvp). MVP is a racially and ethnically diverse veteran cohort that is roughly 20% African American and 7% Hispanic. More than 40,000 of the participants have had prostate cancer, one third of whom are African Americans, giving researchers unprecedented ability to discover factors that impact the development and treatment of the disease in this population. In particular, MVP will provide unique insights into the genetic mutations that drive the development of aggressive prostate cancer in all male veterans, including African Americans. These discoveries will undoubtedly lead to improved screening of and treatment for prostate cancer.
In order to demonstrate clinical utility as well as the infrastructure needs to scale up within the VHA, MVP has launched a pilot project that offers to return clinically actionable genetic results to MVP participants with metastatic prostate cancer, opening the door to new therapies to improve the length and quality of these veterans’ lives. Importantly, the pilot includes cascade testing in family members of enrolled veterans. Given that the original MVP consent did not allow for return of results, and MVP genetic testing is research grade, veterans who volunteer will provide a second consent and undergo clinical genetic testing to confirm the variants. Results from this pilot study also will inform expansion of VA precision oncology efforts for patients with other cancers such as breast cancer or ovarian cancer, where the specific genetic mutations are known to play a role, (eg, BRCA2). In addition, through an interagency agreement with the US Department of Energy (DOE), MVP is leveraging DOE expertise and high-performance computing capabilities to identify clinical and genetic risk factors for prostate cancer that will progress to metastatic disease.
This active research collaboration between POPCaP, MVP, and the Genomic Medicine Service will identify germline BRCA alterations from MVP participants with metastatic prostate cancer and give them access to therapies that may provide better outcomes and access to genetic testing for their family members.
Future Directions
The POPCaP network and its partnership with VA clinical and research efforts is anticipated to provide important insights into barriers and solutions to the implementation of precision oncology for prostate cancer across the VA. These lessons learned may also be relevant for precision oncology care in other settings. As an example, the role of germline testing and genetic counseling is growing more relevant in precision oncology, yet it is clear that the number of men and women dealing with malignancy who actually receive counseling and testing is suboptimal in most health care systems.14 Optimizing the quality and efficiency of oncogenetics within the VA system in a manner that gives access to these services for every veteran in urban or rural environments is an important goal.
The VA has done extensive work in teleoncology and the Genomic Medicine Service provides telehealth genetic counseling service to 90 VA medical facilities nationwide. Expanding on this model to create a distributed network system across the country is an opportunity that will continue to raise VA profile as a leader in this area while providing increased access to genetic services.
Finally, the clinical trials network within POPCaP already has provided valuable insights into how research efforts that originate within the VA can leverage the VA’s strengths. The use of the NPOP centralized sequencing platform to identify potentially targetable alterations across medical centers provides the potential to bring critical access to research to veterans where they live through virtual clinical trials. The VA has a centralized institutional review board that can service large multisite study participation efficiently across the VA. The promise of virtual clinical trials to interrogate relatively rare biomarkers would benefit from institution of a virtual clinical trials workflow. In theory patients with a potentially targetable biomarker could be identified through the centralized DNA sequencing platform and a clinical trial team of virtual investigators and research coordinators would work with health care providers at sites for study startup and performance. Efforts to design and implement this approach are actively being pursued.
The goal of the VA/PCF POPCaP network is to make certain that every veteran has access to appropriate genetic and genomic testing and that the results are utilized so that veterans with targetable alterations receive the best clinical care and have access to clinical trials that could benefit them individually while advancing knowledge that benefits all.
1. Montgomery B, Williams C. Prostate cancer federal health care data trends. https://www.mdedge.com/fedprac/article/208077/oncology/prostate-cancer-federal-health-care-data-trends. Published September 1, 2019. Accessed July 16, 2020.
2. Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer [published correction appears in Cell. 2015 Jul 16;162(2):454]. Cell. 2015;161(5):1215-1228. doi:10.1016/j.cell.2015.05.001
3. Quigley DA, Dang HX, Zhao SG, et al. Genomic hallmarks and structural variation in metastatic prostate cancer [published correction appears in Cell. 2018 Oct 18;175(3):889]. Cell. 2018;174(3):758-769.e9. doi:10.1016/j.cell.2018.06.039
4. Pritchard CC, Offit K, Nelson PS. DNA-repair gene mutations in metastatic prostate cancer. N Engl J Med. 2016;375(18):1804-1805. doi:10.1056/NEJMc1611137
5. Guillem JG. Molecular diagnosis of hereditary nonpolyposis colon cancer. N Engl J Med. 1998;339(13):924-925. doi:10.1056/nejm199809243391316
6. Ryan S, Jenkins MA, Win AK. Risk of prostate cancer in Lynch syndrome: a systematic review and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2014;23(3):437-449. doi:10.1158/1055-9965.EPI-13-1165
7. Abida W, Cheng ML, Armenia J, et al. Analysis of the prevalence of microsatellite instability in prostate cancer and response to immune checkpoint blockade. JAMA Oncol. 2019;5(4):471-478. doi:10.1001/jamaoncol.2018.5801
8. Graham LS, Montgomery B, Cheng HH, et al. Mismatch repair deficiency in metastatic prostate cancer: Response to PD-1 blockade and standard therapies. PLoS One. 2020;15(5):e0233260. Published 2020 May 26. doi:10.1371/journal.pone.0233260
9. Yu HA, Planchard D, Lovly CM. Sequencing therapy for genetically defined subgroups of non-small cell lung cancer. Am Soc Clin Oncol Educ Book. 2018;38:726-739. doi:10.1200/EDBK_201331
10. Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375(5):443-453. doi:10.1056/NEJMoa1603144
11. Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917-921. doi:10.1038/nature03445
12. Abida W, Campbell D, Patnaik A, et al. Preliminary results from the TRITON2 study of rucaparib in patients with DNA damage repair deficiency metastatic, castration resistant prostate cancer: updated analyses. Ann Oncol. 2019;30(suppl 5): v325-v355. doi:10.1093/annonc/mdz248
13. de Bono J, Mateo J, Fizazi K, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382(22):2091-2102. doi:10.1056/NEJMoa1911440
14. Graff JN, Huang GD. Leveraging Veterans Health Administration clinical and research resources to accelerate discovery and testing in precision oncology. Fed Pract. 2020;37(suppl 4):S62-S67. doi: 10.12788/fp.0028
1. Montgomery B, Williams C. Prostate cancer federal health care data trends. https://www.mdedge.com/fedprac/article/208077/oncology/prostate-cancer-federal-health-care-data-trends. Published September 1, 2019. Accessed July 16, 2020.
2. Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer [published correction appears in Cell. 2015 Jul 16;162(2):454]. Cell. 2015;161(5):1215-1228. doi:10.1016/j.cell.2015.05.001
3. Quigley DA, Dang HX, Zhao SG, et al. Genomic hallmarks and structural variation in metastatic prostate cancer [published correction appears in Cell. 2018 Oct 18;175(3):889]. Cell. 2018;174(3):758-769.e9. doi:10.1016/j.cell.2018.06.039
4. Pritchard CC, Offit K, Nelson PS. DNA-repair gene mutations in metastatic prostate cancer. N Engl J Med. 2016;375(18):1804-1805. doi:10.1056/NEJMc1611137
5. Guillem JG. Molecular diagnosis of hereditary nonpolyposis colon cancer. N Engl J Med. 1998;339(13):924-925. doi:10.1056/nejm199809243391316
6. Ryan S, Jenkins MA, Win AK. Risk of prostate cancer in Lynch syndrome: a systematic review and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2014;23(3):437-449. doi:10.1158/1055-9965.EPI-13-1165
7. Abida W, Cheng ML, Armenia J, et al. Analysis of the prevalence of microsatellite instability in prostate cancer and response to immune checkpoint blockade. JAMA Oncol. 2019;5(4):471-478. doi:10.1001/jamaoncol.2018.5801
8. Graham LS, Montgomery B, Cheng HH, et al. Mismatch repair deficiency in metastatic prostate cancer: Response to PD-1 blockade and standard therapies. PLoS One. 2020;15(5):e0233260. Published 2020 May 26. doi:10.1371/journal.pone.0233260
9. Yu HA, Planchard D, Lovly CM. Sequencing therapy for genetically defined subgroups of non-small cell lung cancer. Am Soc Clin Oncol Educ Book. 2018;38:726-739. doi:10.1200/EDBK_201331
10. Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375(5):443-453. doi:10.1056/NEJMoa1603144
11. Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917-921. doi:10.1038/nature03445
12. Abida W, Campbell D, Patnaik A, et al. Preliminary results from the TRITON2 study of rucaparib in patients with DNA damage repair deficiency metastatic, castration resistant prostate cancer: updated analyses. Ann Oncol. 2019;30(suppl 5): v325-v355. doi:10.1093/annonc/mdz248
13. de Bono J, Mateo J, Fizazi K, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382(22):2091-2102. doi:10.1056/NEJMoa1911440
14. Graff JN, Huang GD. Leveraging Veterans Health Administration clinical and research resources to accelerate discovery and testing in precision oncology. Fed Pract. 2020;37(suppl 4):S62-S67. doi: 10.12788/fp.0028