Ms. C, age 45, has a history of generalized anxiety disorder, which has been controlled for the past 6 weeks with extended-release (ER) venlafaxine, 37.5 mg/d. Previous medication trials included fluvoxamine, 300 mg/d, for 2 weeks; paroxetine, 20 mg/d, for 1 week; sertraline, 100 mg/d, for 1 week; and citalopram, 20 mg/d, for 2 weeks. For each trial, Ms. C was unable to tolerate standard doses because of substantial adverse effects; she complained that her anxiety would significantly worsen with each course of treatment. Although the adverse effects would eventually subside with continued treatment, they appeared to be the dose-limiting factor for treatment, even when much lower doses were started.
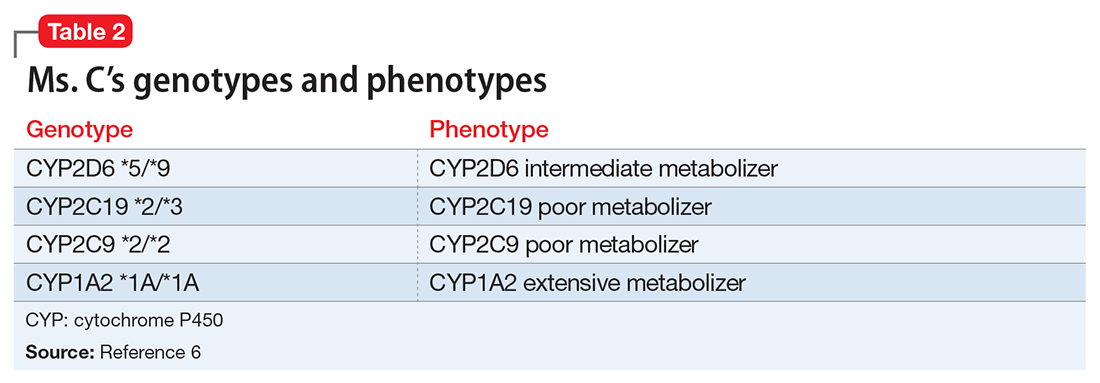
Ms. C’s son recently suggested that she undergo pharmacogenomics testing, and she brings in the results of this testing. The report states that Ms. C has cytochrome P450 (CYP) pharmacogenotypes CYP2D6 *5/*9, CYP2C19 *2/*3, CYP2C9 *2/*2, and CYP1A2 *1A/*1F. Ms. C wants to know if these results explain some of the issues she has had with previous medication trials, and if these results mean that she should be taking a different medication.
The human genome project was a vast, international effort to sequence the entire human genome1 and identify individual differences in drug response, which serves as the basis for pharmacogenomics. Since completion of the human genome project in the early 2000s, the field of pharmacogenomics has advanced, and using pharmacogenomic testing to make therapeutic decisions for medication management is becoming commonplace.2 Although this critical change to how medicine is practiced is exciting, implementation of pharmacogenomics into practice has been varied.2 Therefore, having an understanding of the resources available to guide pharmacogenomics into practice is critical, because the FDA now lists >160 medications that include specific pharmacogenomics information within their package insert.3
CPIC provides guidance for implementing pharmacogenomics
In 2000, the National Institutes of Health established the Pharmacogenomics Knowledge Base (PharmGKB) and the Pharmacogenomics Research Network (PGRN). These 2 resources provide information from cutting-edge research on genomic variation and therapeutic and adverse events, as well as practical implementation of this research.4 As part of their partnership, PharmGKB and PGRN established the Clinical Pharmacogenomics Implementation Consortium (CPIC), which has begun to provide clinical practice guidelines for implementing pharmacogenomic results. Although CPIC does not advocate for pharmacogenomics testing as a standard, it recognizes that this testing is becoming more commonplace, and therefore its guidelines can help clinicians make rational prescribing decisions.4
In a recent partnership among several PGRN members, investigators found that 1 out of 4 pharmacogenomic test results had a potential clinically actionable outcome.2 There are currently >43 gene/drug pairs for which CPIC has provided guidelines; however, >200 other gene/drug pairs are being evaluated.5
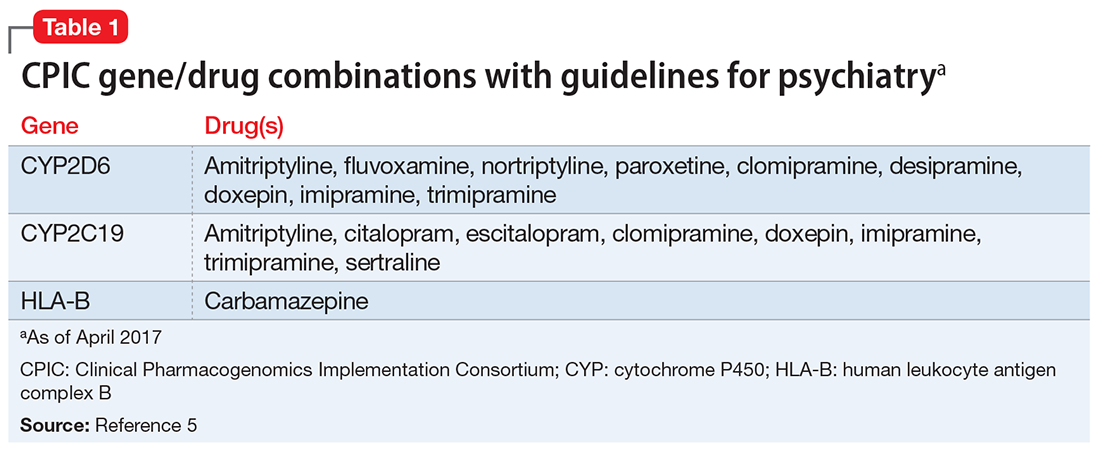
Table 15 lists the current CPIC gene/drug combinations with accompanying published guidelines that are pertinent to psychiatry. For each of these guidelines, experts reviewed the available literature to provide graded therapeutic recommendations: A (“preponderance of evidence is high or moderate in favor of changing prescribing”), B (“preponderance of evidence is weak with little conflicting data”), and C and D (“evidence levels can vary”).4 Looking at the specific genotypes for Ms. C, we can use the information within the CPIC to assign a drug metabolism phenotype for her genotype combinations (Table 2).6
Consider additional resources
In addition to those from the CPIC, guidelines have been developed by other scientific groups, such as the Dutch Pharmacogenetics Working Group and the European Pharmacogenomics Implementation Consortium. Although most of these guidelines are concordant with CPIC, differences exist, which makes it important to be aware of all available resources.