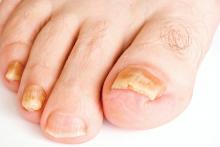
Real-time PCR techniques for identifying the pathogens responsible for onychomycosis can offer some advantages over conventional diagnostic approaches but also have their limitations, say the authors of a study published in Mycoses.
Anissa Z. Hafirassou of Frères-Mentouri, Constantine University, Algeria, and colleagues analyzed nail samples from 70 patients with clinical signs of onychomycosis and 15 healthy controls using four different real-time PCR assays – a panfungal, a pandermatophyte, an assay for Candida and one for Aspergillus – and conventional methods.
The panfungal analysis detected DNA in 57% of cases but the sequencing results were positive in 28% of cases. The sensitivity compared to positive cultures was 47% (Mycoses. 2017 May 16. doi: 10.1111/myc.12629).Most samples were of Trichophyton species and were found in patients with proven onychomycosis. In contrast, the sequencing results from the healthy samples were all negative.
The pandermatophyte analysis found dermatophyte DNA in 60% of cases – most of were proven cases of onychomycosis – representing a sensitivity of 90% compared to positive culture. This analysis showed 90% sensitivity compared to cultures, but there was no correlation between culture results and pandermatophyte RT-PCR in nine cases.
This technique also detected Trichophyton cases in 15 patients who had negative culture results, but found amplification products in three of the control subjects, two of which were Penicillium chrysogenum. However two culture-positive samples showed up as negative with both the panfungal and pandermatophyte methods.
“Due to the low sensitivity of the panfungal assay and the lack of correlation between cultures and PCR results, the possibility of the presence of environmental and colonizing species together with pathological species in nail samples, was studied,” the authors wrote.
Twenty-five fingernail samples that were negative on the panfungal analysis were also tested for Candida and Aspergillus. Candida species were detected in 76% of these samples, and Aspergillus in 60%, while 64% contained mixed populations. Ten samples contained more than one species of Candida and one had two species of Aspergillus.
“Conventional diagnostic methods have several limitations such as time-cost, low sensitivity and the need of skilled personnel,” the authors wrote, noting that the molecular methods also had limitations to their usefulness.
The panfungal method showed low sensitivity, which may have been due to the mix of fungal populations that was found even in healthy controls, the researchers added.
“The pandermatophyte assay was sensitive and specific but only detected dermatophyte species and did not allow differentiation among them,” they wrote.
The role of nondermatophyte species isolated from onychomycosis should be considered carefully, as these are also found in healthy nails, the researchers noted.
The study and one author were supported by the Spanish Fondo de Investigaciones Sanitarias of the Instituto de Salud Carlos III. No conflicts of interest were declared.