User login
2019 Novel Coronavirus: Frequently asked questions for clinicians
The 2019 Novel Coronavirus (2019-nCoV) outbreak has unfolded so rapidly that many clinicians are scrambling to stay on top of it. Here are the answers to some frequently asked questions about how to prepare your clinic to respond to this outbreak.
Keep in mind that the outbreak is moving rapidly. Though scientific and epidemiologic knowledge has increased at unprecedented speed, there is much we don’t know, and some of what we think we know will change. Follow the links for the most up-to-date information.
What should our clinic do first?
Plan ahead with the following:
- Develop a plan for office staff to take travel histories from anyone with a respiratory illness and provide training for those who need it. Travel history at present should include asking about travel to China in the past 14 days, specifically Wuhan city or Hubei province.
- Review up-to-date infection control practices with all office staff and provide training for those who need it.
- Take an inventory of supplies of personal protective equipment (PPE), such as gowns, gloves, masks, eye protection, and N95 respirators or powered air-purifying respirators (PAPRs), and order items that are missing or low in stock.
- Fit-test users of N95 masks for maximal effectiveness.
- Plan where a potential patient would be isolated while obtaining expert advice.
- Know whom to contact at the state or local health department if you have a patient with the appropriate travel history.
The Centers for Disease Control and Prevention has prepared a toolkit to help frontline health care professionals prepare for this virus. Providers need to stay up to date on the latest recommendations, as the situation is changing rapidly.
When should I suspect 2019-nCoV illness, and what should I do?
Take the following steps to assess the concern and respond:
- If a patient with respiratory illness has traveled to China in the past 14 days, immediately put a mask on the patient and move the individual to a private room. Use a negative-pressure room if available.
- Put on appropriate PPE (including gloves, gown, eye protection, and mask) for contact, droplet, and airborne precautions. CDC recommends an N95 respirator mask if available, although we don’t know yet if there is true airborne spread.
- Obtain an accurate travel history, including dates and cities. (Tip: Get the correct spelling, as the English spelling of cities in China can cause confusion.)
- If the patient meets the current CDC definition of “person under investigation” or PUI, or if you need guidance on how to proceed, notify infection control (if you are in a facility that has it) and call your state or local health department immediately.
- Contact public health authorities who can help decide whether the patient should be admitted to airborne isolation or monitored at home with appropriate precautions.
What is the definition of a PUI?
The current definition of a PUI is a person who has fever and symptoms of a respiratory infection (cough, shortness of breath) AND who has EITHER been in Wuhan city or Hubei province in the past 14 days OR had close contact with a person either under investigation for 2019-nCoV infection or with confirmed infection. The definition of a PUI will change over time, so check this link.
How can I test for 2019-nCoV?
As of Jan. 30, 2020, testing is by polymerase chain reaction (PCR) and is available in the United States only through the CDC in Atlanta. Testing should soon be available in state health department laboratories. If public health authorities decide that your patient should be tested, they will instruct you on which samples to obtain.
The full sequence of 2019-nCoV has been shared, so some reference laboratories may develop and validate tests, ideally with assistance from CDC. If testing becomes available, make certain that it is a reputable lab that has carefully validated the test.
Should I test for other viruses?
Because the symptoms of 2019-nCoV infection overlap with those of influenza and other respiratory viruses, PCR testing for other viruses should be considered if it will change management (i.e., change the decision to provide influenza antivirals). Use appropriate PPE while collecting specimens, including eye protection. If 2019-nCoV is a consideration, you may want to send the specimen to a hospital lab for testing, where the sample will be processed under a biosafety hood, rather than doing point-of-care testing in the office.
How dangerous is 2019-nCoV?
The current estimated mortality rate is 2%-3%. That is probably an overestimate, as those with severe disease and those who die are more likely to be tested and reported early in an epidemic.
Our current knowledge is based on preliminary reports from hospitalized patients and will probably change. From the speed of spread and a single family cluster, it seems likely that there are milder cases and perhaps asymptomatic infection.
What else do I need to know about coronaviruses?
Coronaviruses are a large and diverse group of viruses, many of which are animal viruses. Before the discovery of the 2019-nCoV, six coronaviruses were known to infect humans. Four of these (HKU1, NL63, OC43, and 229E) predominantly caused mild to moderate upper respiratory illness, and they are thought to be responsible for 10%-30% of colds. They occasionally cause viral pneumonia and can be detected by some commercial multiplex panels.
Two other coronaviruses have caused outbreaks of severe respiratory illness in people: SARS, which emerged in Southern China in 2002, and MERS in the Middle East, in 2012. Unlike SARS, sporadic cases of MERS continue to occur.
The current outbreak is caused by 2019-nCoV, a previously unknown beta coronavirus. It is most closely related (~96%) to a bat virus and shares about 80% sequence homology with SARS CoV.
Andrew T. Pavia, MD, is the George and Esther Gross Presidential Professor and chief of the division of pediatric infectious disease in the department of pediatrics at the University of Utah, Salt Lake City. He is also director of hospital epidemiology and associate director of antimicrobial stewardship at Primary Children’s Hospital, Salt Lake City. Dr. Pavia has disclosed that he has served as a consultant for Genentech, Merck, and Seqirus and that he has served as associate editor for The Sanford Guide.
This article first appeared on Medscape.com.
The 2019 Novel Coronavirus (2019-nCoV) outbreak has unfolded so rapidly that many clinicians are scrambling to stay on top of it. Here are the answers to some frequently asked questions about how to prepare your clinic to respond to this outbreak.
Keep in mind that the outbreak is moving rapidly. Though scientific and epidemiologic knowledge has increased at unprecedented speed, there is much we don’t know, and some of what we think we know will change. Follow the links for the most up-to-date information.
What should our clinic do first?
Plan ahead with the following:
- Develop a plan for office staff to take travel histories from anyone with a respiratory illness and provide training for those who need it. Travel history at present should include asking about travel to China in the past 14 days, specifically Wuhan city or Hubei province.
- Review up-to-date infection control practices with all office staff and provide training for those who need it.
- Take an inventory of supplies of personal protective equipment (PPE), such as gowns, gloves, masks, eye protection, and N95 respirators or powered air-purifying respirators (PAPRs), and order items that are missing or low in stock.
- Fit-test users of N95 masks for maximal effectiveness.
- Plan where a potential patient would be isolated while obtaining expert advice.
- Know whom to contact at the state or local health department if you have a patient with the appropriate travel history.
The Centers for Disease Control and Prevention has prepared a toolkit to help frontline health care professionals prepare for this virus. Providers need to stay up to date on the latest recommendations, as the situation is changing rapidly.
When should I suspect 2019-nCoV illness, and what should I do?
Take the following steps to assess the concern and respond:
- If a patient with respiratory illness has traveled to China in the past 14 days, immediately put a mask on the patient and move the individual to a private room. Use a negative-pressure room if available.
- Put on appropriate PPE (including gloves, gown, eye protection, and mask) for contact, droplet, and airborne precautions. CDC recommends an N95 respirator mask if available, although we don’t know yet if there is true airborne spread.
- Obtain an accurate travel history, including dates and cities. (Tip: Get the correct spelling, as the English spelling of cities in China can cause confusion.)
- If the patient meets the current CDC definition of “person under investigation” or PUI, or if you need guidance on how to proceed, notify infection control (if you are in a facility that has it) and call your state or local health department immediately.
- Contact public health authorities who can help decide whether the patient should be admitted to airborne isolation or monitored at home with appropriate precautions.
What is the definition of a PUI?
The current definition of a PUI is a person who has fever and symptoms of a respiratory infection (cough, shortness of breath) AND who has EITHER been in Wuhan city or Hubei province in the past 14 days OR had close contact with a person either under investigation for 2019-nCoV infection or with confirmed infection. The definition of a PUI will change over time, so check this link.
How can I test for 2019-nCoV?
As of Jan. 30, 2020, testing is by polymerase chain reaction (PCR) and is available in the United States only through the CDC in Atlanta. Testing should soon be available in state health department laboratories. If public health authorities decide that your patient should be tested, they will instruct you on which samples to obtain.
The full sequence of 2019-nCoV has been shared, so some reference laboratories may develop and validate tests, ideally with assistance from CDC. If testing becomes available, make certain that it is a reputable lab that has carefully validated the test.
Should I test for other viruses?
Because the symptoms of 2019-nCoV infection overlap with those of influenza and other respiratory viruses, PCR testing for other viruses should be considered if it will change management (i.e., change the decision to provide influenza antivirals). Use appropriate PPE while collecting specimens, including eye protection. If 2019-nCoV is a consideration, you may want to send the specimen to a hospital lab for testing, where the sample will be processed under a biosafety hood, rather than doing point-of-care testing in the office.
How dangerous is 2019-nCoV?
The current estimated mortality rate is 2%-3%. That is probably an overestimate, as those with severe disease and those who die are more likely to be tested and reported early in an epidemic.
Our current knowledge is based on preliminary reports from hospitalized patients and will probably change. From the speed of spread and a single family cluster, it seems likely that there are milder cases and perhaps asymptomatic infection.
What else do I need to know about coronaviruses?
Coronaviruses are a large and diverse group of viruses, many of which are animal viruses. Before the discovery of the 2019-nCoV, six coronaviruses were known to infect humans. Four of these (HKU1, NL63, OC43, and 229E) predominantly caused mild to moderate upper respiratory illness, and they are thought to be responsible for 10%-30% of colds. They occasionally cause viral pneumonia and can be detected by some commercial multiplex panels.
Two other coronaviruses have caused outbreaks of severe respiratory illness in people: SARS, which emerged in Southern China in 2002, and MERS in the Middle East, in 2012. Unlike SARS, sporadic cases of MERS continue to occur.
The current outbreak is caused by 2019-nCoV, a previously unknown beta coronavirus. It is most closely related (~96%) to a bat virus and shares about 80% sequence homology with SARS CoV.
Andrew T. Pavia, MD, is the George and Esther Gross Presidential Professor and chief of the division of pediatric infectious disease in the department of pediatrics at the University of Utah, Salt Lake City. He is also director of hospital epidemiology and associate director of antimicrobial stewardship at Primary Children’s Hospital, Salt Lake City. Dr. Pavia has disclosed that he has served as a consultant for Genentech, Merck, and Seqirus and that he has served as associate editor for The Sanford Guide.
This article first appeared on Medscape.com.
The 2019 Novel Coronavirus (2019-nCoV) outbreak has unfolded so rapidly that many clinicians are scrambling to stay on top of it. Here are the answers to some frequently asked questions about how to prepare your clinic to respond to this outbreak.
Keep in mind that the outbreak is moving rapidly. Though scientific and epidemiologic knowledge has increased at unprecedented speed, there is much we don’t know, and some of what we think we know will change. Follow the links for the most up-to-date information.
What should our clinic do first?
Plan ahead with the following:
- Develop a plan for office staff to take travel histories from anyone with a respiratory illness and provide training for those who need it. Travel history at present should include asking about travel to China in the past 14 days, specifically Wuhan city or Hubei province.
- Review up-to-date infection control practices with all office staff and provide training for those who need it.
- Take an inventory of supplies of personal protective equipment (PPE), such as gowns, gloves, masks, eye protection, and N95 respirators or powered air-purifying respirators (PAPRs), and order items that are missing or low in stock.
- Fit-test users of N95 masks for maximal effectiveness.
- Plan where a potential patient would be isolated while obtaining expert advice.
- Know whom to contact at the state or local health department if you have a patient with the appropriate travel history.
The Centers for Disease Control and Prevention has prepared a toolkit to help frontline health care professionals prepare for this virus. Providers need to stay up to date on the latest recommendations, as the situation is changing rapidly.
When should I suspect 2019-nCoV illness, and what should I do?
Take the following steps to assess the concern and respond:
- If a patient with respiratory illness has traveled to China in the past 14 days, immediately put a mask on the patient and move the individual to a private room. Use a negative-pressure room if available.
- Put on appropriate PPE (including gloves, gown, eye protection, and mask) for contact, droplet, and airborne precautions. CDC recommends an N95 respirator mask if available, although we don’t know yet if there is true airborne spread.
- Obtain an accurate travel history, including dates and cities. (Tip: Get the correct spelling, as the English spelling of cities in China can cause confusion.)
- If the patient meets the current CDC definition of “person under investigation” or PUI, or if you need guidance on how to proceed, notify infection control (if you are in a facility that has it) and call your state or local health department immediately.
- Contact public health authorities who can help decide whether the patient should be admitted to airborne isolation or monitored at home with appropriate precautions.
What is the definition of a PUI?
The current definition of a PUI is a person who has fever and symptoms of a respiratory infection (cough, shortness of breath) AND who has EITHER been in Wuhan city or Hubei province in the past 14 days OR had close contact with a person either under investigation for 2019-nCoV infection or with confirmed infection. The definition of a PUI will change over time, so check this link.
How can I test for 2019-nCoV?
As of Jan. 30, 2020, testing is by polymerase chain reaction (PCR) and is available in the United States only through the CDC in Atlanta. Testing should soon be available in state health department laboratories. If public health authorities decide that your patient should be tested, they will instruct you on which samples to obtain.
The full sequence of 2019-nCoV has been shared, so some reference laboratories may develop and validate tests, ideally with assistance from CDC. If testing becomes available, make certain that it is a reputable lab that has carefully validated the test.
Should I test for other viruses?
Because the symptoms of 2019-nCoV infection overlap with those of influenza and other respiratory viruses, PCR testing for other viruses should be considered if it will change management (i.e., change the decision to provide influenza antivirals). Use appropriate PPE while collecting specimens, including eye protection. If 2019-nCoV is a consideration, you may want to send the specimen to a hospital lab for testing, where the sample will be processed under a biosafety hood, rather than doing point-of-care testing in the office.
How dangerous is 2019-nCoV?
The current estimated mortality rate is 2%-3%. That is probably an overestimate, as those with severe disease and those who die are more likely to be tested and reported early in an epidemic.
Our current knowledge is based on preliminary reports from hospitalized patients and will probably change. From the speed of spread and a single family cluster, it seems likely that there are milder cases and perhaps asymptomatic infection.
What else do I need to know about coronaviruses?
Coronaviruses are a large and diverse group of viruses, many of which are animal viruses. Before the discovery of the 2019-nCoV, six coronaviruses were known to infect humans. Four of these (HKU1, NL63, OC43, and 229E) predominantly caused mild to moderate upper respiratory illness, and they are thought to be responsible for 10%-30% of colds. They occasionally cause viral pneumonia and can be detected by some commercial multiplex panels.
Two other coronaviruses have caused outbreaks of severe respiratory illness in people: SARS, which emerged in Southern China in 2002, and MERS in the Middle East, in 2012. Unlike SARS, sporadic cases of MERS continue to occur.
The current outbreak is caused by 2019-nCoV, a previously unknown beta coronavirus. It is most closely related (~96%) to a bat virus and shares about 80% sequence homology with SARS CoV.
Andrew T. Pavia, MD, is the George and Esther Gross Presidential Professor and chief of the division of pediatric infectious disease in the department of pediatrics at the University of Utah, Salt Lake City. He is also director of hospital epidemiology and associate director of antimicrobial stewardship at Primary Children’s Hospital, Salt Lake City. Dr. Pavia has disclosed that he has served as a consultant for Genentech, Merck, and Seqirus and that he has served as associate editor for The Sanford Guide.
This article first appeared on Medscape.com.
Prevalence of Staphylococcus aureus and Use of Antistaphylococcal Therapy in Children Hospitalized with Pneumonia
Although Staphylococcus aureus pneumonia is common in children with cystic fibrosis and those with healthcare-associated infections (eg, ventilator-associated pneumonia),1,2 S. aureus is an uncommon cause of community-acquired pneumonia in children. In recent years, concerns have arisen about the increasing frequency and severity of staphylococcal pneumonia, largely fueled by the emergence of community-associated methicillin-resistant S. aureus (MRSA).3,4 Thus, therapy with clindamycin or vancomycin, both active against MRSA, has been recommended when S. aureus is suspected.5 Given the lack of rapid and sensitive approaches to the detection of the etiologies of pneumonia, antibiotic selection is most often empirical, contributing to overuse of anti-MRSA antibiotics. In addition, resistance against these antibiotics, especially clindamycin, has been increasing.6,7
A better understanding of the likelihood of staphylococcal pneumonia would help to optimize empirical antibiotic selection, allowing for judicious use of antistaphylococcal antibiotics, while also avoiding poor outcomes due to delays in effective treatment when S. aureus is present.8 Using data from a multicenter, population-based study of pneumonia hospitalizations in children, we sought to describe the prevalence, clinical characteristics, and in-hospital outcomes of staphylococcal pneumonia and the prevalence of antistaphylococcal antibiotic use.
METHODS
The Etiology of Pneumonia in the Community (EPIC) study was a prospective, active, population-based surveillance study of pneumonia hospitalizations among children (age <18 years) conducted between 2010 and 2012 at three children’s hospitals, including two in Tennessee and one in Utah.9 Children hospitalized with clinical evidence of pneumonia and radiographic evidence confirmed by a blinded review by study radiologists were enrolled. Etiologic assessments included blood analysis for bacterial culture, serology for eight respiratory viruses, pneumococcal and group A streptococcal polymerase chain reaction (PCR), and naso/oro-pharyngeal swabs for PCR for 13 respiratory viruses, Mycoplasma pneumoniae, and Chlamydophila pneumoniae. Data from other clinical specimens (pleural fluid, high-quality endotracheal aspirate, or quantified bronchoalveolar lavage fluid) were also recorded. For this study, we included only children with at least one bacterial culture and complete information about antibiotic use. Those with confirmed fungal pneumonia were excluded. Additional details regarding the study population and methods have been published previously.9
Staphylococcal pneumonia was defined based on the detection of S. aureus by culture (any site) or PCR (pleural fluid only), regardless of codetection of other pathogens. Antibiotic susceptibility profiles were used to classify S. aureus isolates as MRSA or methicillin-sensitive S. aureus (MSSA). The remaining children were classified as nonstaphylococcal pneumonia including children with other bacterial pathogens detected (classified as other bacterial pneumonia, excludes atypical bacteria), atypical bacteria, viruses, and no pathogens detected.
Use of anti-MRSA antibiotics (vancomycin, clindamycin, linezolid, doxycycline, and trimethoprim-sulfamethoxazole) and any antistaphylococcal antibiotics (anti-MRSA agents plus oxacillin, nafcillin, and cefazolin) during and after the first two calendar days of admission was identified by medical record review.
Descriptive statistics included number (%) and median (interquartile range, [IQR]) for categorical and continuous variables, respectively. Baseline clinical characteristics and outcomes were compared between children with staphylococcal versus nonstaphylococcal pneumonia, those with staphylococcal versus other bacterial pneumonia, and those with MRSA versus MSSA pneumonia using Wilcoxon rank-sum and Pearson’s chi-square tests where appropriate. To account for multiple comparisons, we used a Bonferroni corrected P value threshold of <.001 to determine statistical significance.
RESULTS
Of the 2,358 children enrolled in the EPIC study hospitalized with radiographically confirmed pneumonia, 2,146 (91.0%) had ≥1 bacterial culture obtained. Two children with Histoplasma capsulatum fungal infection and six children with incomplete antibiotic utilization data were excluded, yielding a final study population of 2,138 children. Among these, blood samples were obtained from 2,134 (>99%) children for culture, pleural fluid from 87 (4%) children, bronchoalveolar lavage fluid from 31 (1%) children, and endotracheal aspirate from 80 (4%) children. Across all culture types, there were 2,332 initial cultures; 2,150 (92%) were collected within the first 24 hours.
Staphylococcal pneumonia was detected in 23 of the 2,138 children (1% [95% CI 0.7, 1.6]; 17 MRSA, 6 MSSA). Of these, 6/23 (26%) had bacteremia, 12/23 (52%) had a positive pleural fluid, and 9/23 (39%) had a positive culture from bronchoalveolar lavage fluid or endotracheal aspirate; 4/23 (17%) children had S. aureus detected from more than one site. Three children (13%) with S. aureus had a viral codetection, including two with influenza.
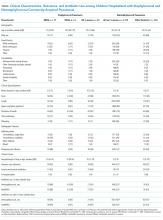
Compared with children with nonstaphylococcal pneumonia, those with staphylococcal pneumonia were more likely to have a parapneumonic effusion (78% vs 12%, P < .001), but less likely to have cough (78% vs 95%, P < .001). Other baseline characteristics were similar between the two groups. Children with staphylococcal pneumonia had more adverse outcomes than those without (Table), including longer median length of stay (10 vs 3 days, P < .001), more frequent admission to intensive care (83% vs 21%, P < .001), and more frequent invasive mechanical ventilation (65% vs 7%, P < .001). Similar findings were noted when staphylococcal pneumonia was compared with pneumonia caused due to other bacterial pathogens (n = 124). There were no significant differences in baseline characteristics or clinical course between children with MRSA and MSSA pneumonia, although the numbers were small. Overall, S. aureus was detected in 18/267 (7%) children with parapneumonic effusion and 19/462 (4%) children admitted to intensive care. Importantly, there were no confirmed S. aureus cases among children with less severe pneumonia, defined as lacking both parapneumonic effusion and intensive care admission (n = 1,488).
Overall, 519 children (24%) received antistaphylococcal therapy during their hospitalization (512/519, 99% received anti-MRSA therapy), including 22 of the 23 children with S. aureus detected (the only child without antistaphylococcal therapy had S. aureus detected from a high-quality endotracheal tube aspirate only and also had respiratory syncytial virus detected). Clindamycin was most often used (n = 266, 51%), followed by vancomycin (n = 128, 24%), clindamycin plus vancomycin (n = 83, 16%), and others (n = 42, 8%). During the first two days of hospitalization, 479 children (22%) received antistaphylococcal therapy (477 received anti-MRSA therapy). After the first two days, 351 children (16%) received antistaphylococcal therapy (346/351, 99% received anti-MRSA therapy). Use of antistaphylococcal therapy was very common in those admitted to intensive care (182/462, 39%; all but two received anti-MRSA therapy) and in those requiring invasive mechanical ventilation (103/159, 65%). Among those lacking both parapneumonic effusion and intensive care admission (n = 1488), 232 (16%) received antistaphylococcal therapy.
DISCUSSION
In our large, population-based study of >2,000 children hospitalized with community-acquired pneumonia, S. aureus was identified in only 1% of children. Compared with children with other pneumonia etiologies, staphylococcal pneumonia was associated with increased disease severity. Among the small numbers studied, no differences in outcomes were found between children with MRSA and MSSA disease. Despite the low prevalence of staphylococcal pneumonia, almost 1 in 4 children received antistaphylococcal antibiotic therapy; anti-MRSA therapy was used almost exclusively.
The severity of staphylococcal pneumonia was striking, with >80% of children with S. aureus detected being admitted to intensive care, about 65% requiring invasive mechanical ventilation, and >75% with parapneumonic effusion. These findings are similar to those of prior retrospective studies.4,10 The association between staphylococcal pneumonia and adverse outcomes underscores the importance of prompt institution of antimicrobial therapy targeting S. aureus in high-risk patients. This is noteworthy given recent epidemiological data demonstrating increases in MSSA relative to MRSA infections in children,6 and the known superiority of beta-lactam versus vancomycin for MSSA infections, including pneumonia.11
Although detection of staphylococcal infection was rare, almost a quarter of children received antistaphylococcal therapy; nearly all of these children received anti-MRSA therapy. Confirming a bacterial etiology of pneumonia, however, is challenging. Given the severity associated with staphylococcal pneumonia, it is not surprising that use of antistaphylococcal therapy outpaced staphylococcal detections. Antistaphylococcal therapy was especially common in those with severe pneumonia, suggesting that disease severity is an important factor that influences initial antibiotic treatment decisions. Even so, two children with MRSA detected did not initially receive anti-MRSA therapy, highlighting the challenge of balancing judicious antibiotic selection along with ensuring effective treatment. Perhaps more striking is the finding that 16% of children received antistaphylococcal therapy beyond the first two days of hospitalization, presumably after the initial culture results were available. This suggests that clinicians are reluctant to stop antistaphylococcal therapy when the etiology is unknown, although certain features, such as negative cultures, rapid clinical improvement, and lack of risk factors for staphylococcal disease, may provide important clues to support de-escalation of empiric antibiotic therapy. It is also possible that some antibiotics with antistaphylococcal activity were used for alternative indications (eg, clindamycin for penicillin allergy or concern for aspiration pneumonia).
A simple strategy for tailoring antibiotic treatment is maximizing opportunities to identify a causative pathogen. Despite the very low yield of blood cultures in children with pneumonia overall, bacteremia is more common in children with severe pneumonia and those with parapneumonic effusion, especially when cultures are obtained prior to antibiotic use.12,13 Similarly, obtaining pleural fluid is often therapeutic and significantly improves the chances of identifying a bacterial pathogen.14 Moreover, at least one study suggests that S. aureus is much less likely in cases of culture-negative parapneumonic effusions.15 Institutional guidelines, order sets, and antimicrobial stewardship teams are also effective strategies that can facilitate judicious antibiotic use. In particular, stewardship experts can be very useful in assisting clinicians around de-escalation of therapy.16 Use of procalcitonin, a biomarker associated with bacterial infections,17 and prognostic tools to identify risk for adverse outcomes,18 may also inform treatment decisions and are deserving of further study.
Our study must be considered in
Our study demonstrates a very low prevalence of S. aureus detection among children hospitalized with pneumonia and highlights the association between staphylococcal disease and adverse in-hospital outcomes. We also document important discrepancies between disease prevalence and utilization of antistaphylococcal therapy, especially anti-MRSA therapy. Improved approaches are needed to minimize overuse of antistaphylococcal antibiotics while also ensuring adequate therapy for those who need it.
Disclosures
Drs. Zhu, Edwards, Self, Ampofo, Arnold, McCullers, and Williams report grants from the Centers for Disease Control and Prevention during the conduct of the study. Ms. Frush has nothing to disclose. Dr. Jain has nothing to disclose. Dr. Grijalva reports other from Merck, grants and other from Sanofi, other from Pfizer, grants from CDC, grants from AHRQ, grants from NIH, and grants from Campbell Alliance, outside the submitted work. Dr. Self reports grants from CDC, during the conduct of the study; personal fees from Cempra Pharmaceuticals, grants and personal fees from Ferring Pharmaceuticals, personal fees from BioTest AG, personal fees from Abbott Point of Care, personal fees from Gilead Pharmaceuticals, personal fees from Pfizer, grants from Merck, outside the submitted work. Dr. Thomsen has nothing to disclose. Dr. Ampofo reports grants from CDC, during the conduct of the study; other from GlaxoSmithKline, other from Cubist Pharmaceuticals outside the submitted work; and KA collaborate with BioFire Diagnostics, Inc. (formerly Idaho Technology, Inc.) on several NIH grants. Dr. Pavia reports grants from NAID/NIH, grants from NAID/NIH, grants from CDC, personal fees from WebMD, personal fees from Antimicrobial Therapy Inc., outside the submitted work.
Funding
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number K23AI104779 to D.J.W. and Award 1K23AI113150 to I.P.T., the National Institute of General Medical Sciences under Award K23GM110469 to W.H.S., and the Agency for Healthcare Research and Quality under Award R03HS022342 to C.G.G. The EPIC study was supported by the Influenza Division in the National Center for Immunizations and Respiratory Diseases at the Centers for Disease Control and Prevention through cooperative agreements with each study site and was based on a competitive research funding opportunity. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Institute of Allergy and Infectious Diseases, the National Institute of General Medical Sciences, the Agency for Healthcare Research and Quality, or the Centers for Disease Control and Prevention.
1. Akil N, Muhlebach MS. Biology and management of methicillin resistant Staphylococcus aureus in cystic fibrosis. Pediatr Pulmonol. 2018. doi: 10.1002/ppul.24139. PubMed
2. Srinivasan R, Asselin J, Gildengorin G, Wiener-Kronish J, Flori HR. A prospective study of ventilator-associated pneumonia in children. Pediatrics.
2009;123(4):1108-1115. doi: 10.1542/peds.2008-1211. PubMed
3. Gonzalez BE, Martinez-Aguilar G, Hulten KG, et al. Severe Staphylococcal sepsis in adolescents in the era of community-acquired methicillin-resistant Staphylococcus aureus. Pediatrics. 2005;115(3):642-648. doi: 10.1542/peds.2004-2300. PubMed
4. Carrillo-Marquez MA, Hulten KG, Hammerman W, Lamberth L, Mason EO, Kaplan SL. Staphylococcus aureus pneumonia in children in the era of community-acquired methicillin-resistance at Texas Children’s Hospital. Pediatr Infect Dis J. 2011;30(7):545-550. doi: 10.1097/INF.0b013e31821618be. PubMed
5. Bradley JS, Byington CL, Shah SS, et al. The management of community-acquired pneumonia in infants and children older than 3 months of age: clinical practice guidelines by the pediatric infectious diseases society and the infectious diseases society of America. Clin Infect Dis. 2011;53(7):e25-e76. doi: 10.1093/cid/cir531. PubMed
6. Sutter DE, Milburn E, Chukwuma U, Dzialowy N, Maranich AM, Hospenthal DR. Changing susceptibility of Staphylococcus aureus in a US pediatric population. Pediatrics. 2016;137(4):e20153099–e20153099. doi: 10.1542/peds.2015-3099. PubMed
7. Sakoulas G, Moellering RC, Jr. Increasing antibiotic resistance among methicillin-resistant Staphylococcus aureus strains. Clin Infect Dis. 2008;46(Suppl 5):S360-S367. doi: 10.1086/533592. PubMed
8. Rubinstein E, Kollef MH, Nathwani D. Pneumonia caused by methicillin-resistant
Staphylococcus aureus. Clin Infect Dis. 2008;46(Suppl 5):S378-S385. doi: 10.1086/533594. PubMed
9. Jain S, Williams DJ, Arnold SR, et al. Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med. 2015;372(9):835-845. doi: 10.1056/NEJMoa1405870. PubMed
10. Kallen AJ, Reed C, Patton M, Arnold KE, Finelli L, Hageman J. Staphylococcus aureus community-onset pneumonia in patients admitted to children’s hospitals during autumn and winter of 2006-2007. Epidemiol Infect. 2010;138(5):666-672. doi: 10.1017/S095026880999135X. PubMed
11. González C, Rubio M, Romero-Vivas J, González M, Picazo JJ. Bacteremic pneumonia due to Staphylococcus aureus: A comparison of disease caused by methicillin-resistant and methicillin-susceptible organisms. Clin Infect Dis. 1999;29(5):1171-1177. doi: 10.1086/313440. PubMed
12. Myers AL, Hall M, Williams DJ, et al. Prevalence of bacteremia in hospitalized pediatric patients with community-acquired pneumonia. Pediatr Infect Dis J. 2013;32(7):736-740. doi: 10.1097/INF.0b013e318290bf63. PubMed
13. Iroh Tam PY, Bernstein E, Ma X, Ferrieri P. Blood culture in evaluation of pediatric community-acquired pneumonia: A systematic review and meta-analysis. Hosp Pediatr. 2015;5(6):324-336. doi: 10.1542/hpeds.2014-0138. PubMed
14. Byington CL, Spencer LY, Johnson TA, et al. An epidemiological investigation of a sustained high rate of pediatric parapneumonic empyema: risk factors and microbiological associations. Clin Infect Dis. 2002;34(4):434-440. doi: 10.1086/338460. PubMed
15. Blaschke AJ, Heyrend C, Byington CL, et al. Molecular analysis improves pathogen identifi cation and epidemiologic study of pediatric parapneumonic empyema. Pediatr Infect Dis J. 2011;30(4):289-294. doi: 10.1097/INF.0b013e3182002d14. PubMed
16. Banerjee R, Teng CB, Cunningham SA, et al. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identifi cation and susceptibility testing. Clin Infect Dis. 2015;61(7):1071-1080. doi: 10.1093/cid/civ447. PubMed
17. Stockmann C, Ampofo K, Killpack J, et al. Procalcitonin accurately identifies hospitalized children with low risk of bacterial community-acquired pneumonia. J Pediatr Infect Dis Soc. 2018;7(1):46–53. doi: 10.1093/jpids/piw091. PubMed
18. Williams DJ, Zhu Y, Grijalva CG, et al. Predicting severe pneumonia outcomes in children. Pediatrics. 2016;138(4). doi: 10.1542/peds.2016-1019. PubMed
19. Brogan TV, Hall M, Williams DJ, et al. Variability in processes of care and outcomes among children hospitalized with community-acquired pneumonia. Pediatr Infect Dis J. 2012;31(10):1036-1041. doi: 10.1097/INF.0b013e-31825f2b10. PubMed
Although Staphylococcus aureus pneumonia is common in children with cystic fibrosis and those with healthcare-associated infections (eg, ventilator-associated pneumonia),1,2 S. aureus is an uncommon cause of community-acquired pneumonia in children. In recent years, concerns have arisen about the increasing frequency and severity of staphylococcal pneumonia, largely fueled by the emergence of community-associated methicillin-resistant S. aureus (MRSA).3,4 Thus, therapy with clindamycin or vancomycin, both active against MRSA, has been recommended when S. aureus is suspected.5 Given the lack of rapid and sensitive approaches to the detection of the etiologies of pneumonia, antibiotic selection is most often empirical, contributing to overuse of anti-MRSA antibiotics. In addition, resistance against these antibiotics, especially clindamycin, has been increasing.6,7
A better understanding of the likelihood of staphylococcal pneumonia would help to optimize empirical antibiotic selection, allowing for judicious use of antistaphylococcal antibiotics, while also avoiding poor outcomes due to delays in effective treatment when S. aureus is present.8 Using data from a multicenter, population-based study of pneumonia hospitalizations in children, we sought to describe the prevalence, clinical characteristics, and in-hospital outcomes of staphylococcal pneumonia and the prevalence of antistaphylococcal antibiotic use.
METHODS
The Etiology of Pneumonia in the Community (EPIC) study was a prospective, active, population-based surveillance study of pneumonia hospitalizations among children (age <18 years) conducted between 2010 and 2012 at three children’s hospitals, including two in Tennessee and one in Utah.9 Children hospitalized with clinical evidence of pneumonia and radiographic evidence confirmed by a blinded review by study radiologists were enrolled. Etiologic assessments included blood analysis for bacterial culture, serology for eight respiratory viruses, pneumococcal and group A streptococcal polymerase chain reaction (PCR), and naso/oro-pharyngeal swabs for PCR for 13 respiratory viruses, Mycoplasma pneumoniae, and Chlamydophila pneumoniae. Data from other clinical specimens (pleural fluid, high-quality endotracheal aspirate, or quantified bronchoalveolar lavage fluid) were also recorded. For this study, we included only children with at least one bacterial culture and complete information about antibiotic use. Those with confirmed fungal pneumonia were excluded. Additional details regarding the study population and methods have been published previously.9
Staphylococcal pneumonia was defined based on the detection of S. aureus by culture (any site) or PCR (pleural fluid only), regardless of codetection of other pathogens. Antibiotic susceptibility profiles were used to classify S. aureus isolates as MRSA or methicillin-sensitive S. aureus (MSSA). The remaining children were classified as nonstaphylococcal pneumonia including children with other bacterial pathogens detected (classified as other bacterial pneumonia, excludes atypical bacteria), atypical bacteria, viruses, and no pathogens detected.
Use of anti-MRSA antibiotics (vancomycin, clindamycin, linezolid, doxycycline, and trimethoprim-sulfamethoxazole) and any antistaphylococcal antibiotics (anti-MRSA agents plus oxacillin, nafcillin, and cefazolin) during and after the first two calendar days of admission was identified by medical record review.
Descriptive statistics included number (%) and median (interquartile range, [IQR]) for categorical and continuous variables, respectively. Baseline clinical characteristics and outcomes were compared between children with staphylococcal versus nonstaphylococcal pneumonia, those with staphylococcal versus other bacterial pneumonia, and those with MRSA versus MSSA pneumonia using Wilcoxon rank-sum and Pearson’s chi-square tests where appropriate. To account for multiple comparisons, we used a Bonferroni corrected P value threshold of <.001 to determine statistical significance.
RESULTS
Of the 2,358 children enrolled in the EPIC study hospitalized with radiographically confirmed pneumonia, 2,146 (91.0%) had ≥1 bacterial culture obtained. Two children with Histoplasma capsulatum fungal infection and six children with incomplete antibiotic utilization data were excluded, yielding a final study population of 2,138 children. Among these, blood samples were obtained from 2,134 (>99%) children for culture, pleural fluid from 87 (4%) children, bronchoalveolar lavage fluid from 31 (1%) children, and endotracheal aspirate from 80 (4%) children. Across all culture types, there were 2,332 initial cultures; 2,150 (92%) were collected within the first 24 hours.
Staphylococcal pneumonia was detected in 23 of the 2,138 children (1% [95% CI 0.7, 1.6]; 17 MRSA, 6 MSSA). Of these, 6/23 (26%) had bacteremia, 12/23 (52%) had a positive pleural fluid, and 9/23 (39%) had a positive culture from bronchoalveolar lavage fluid or endotracheal aspirate; 4/23 (17%) children had S. aureus detected from more than one site. Three children (13%) with S. aureus had a viral codetection, including two with influenza.
Compared with children with nonstaphylococcal pneumonia, those with staphylococcal pneumonia were more likely to have a parapneumonic effusion (78% vs 12%, P < .001), but less likely to have cough (78% vs 95%, P < .001). Other baseline characteristics were similar between the two groups. Children with staphylococcal pneumonia had more adverse outcomes than those without (Table), including longer median length of stay (10 vs 3 days, P < .001), more frequent admission to intensive care (83% vs 21%, P < .001), and more frequent invasive mechanical ventilation (65% vs 7%, P < .001). Similar findings were noted when staphylococcal pneumonia was compared with pneumonia caused due to other bacterial pathogens (n = 124). There were no significant differences in baseline characteristics or clinical course between children with MRSA and MSSA pneumonia, although the numbers were small. Overall, S. aureus was detected in 18/267 (7%) children with parapneumonic effusion and 19/462 (4%) children admitted to intensive care. Importantly, there were no confirmed S. aureus cases among children with less severe pneumonia, defined as lacking both parapneumonic effusion and intensive care admission (n = 1,488).
Overall, 519 children (24%) received antistaphylococcal therapy during their hospitalization (512/519, 99% received anti-MRSA therapy), including 22 of the 23 children with S. aureus detected (the only child without antistaphylococcal therapy had S. aureus detected from a high-quality endotracheal tube aspirate only and also had respiratory syncytial virus detected). Clindamycin was most often used (n = 266, 51%), followed by vancomycin (n = 128, 24%), clindamycin plus vancomycin (n = 83, 16%), and others (n = 42, 8%). During the first two days of hospitalization, 479 children (22%) received antistaphylococcal therapy (477 received anti-MRSA therapy). After the first two days, 351 children (16%) received antistaphylococcal therapy (346/351, 99% received anti-MRSA therapy). Use of antistaphylococcal therapy was very common in those admitted to intensive care (182/462, 39%; all but two received anti-MRSA therapy) and in those requiring invasive mechanical ventilation (103/159, 65%). Among those lacking both parapneumonic effusion and intensive care admission (n = 1488), 232 (16%) received antistaphylococcal therapy.
DISCUSSION
In our large, population-based study of >2,000 children hospitalized with community-acquired pneumonia, S. aureus was identified in only 1% of children. Compared with children with other pneumonia etiologies, staphylococcal pneumonia was associated with increased disease severity. Among the small numbers studied, no differences in outcomes were found between children with MRSA and MSSA disease. Despite the low prevalence of staphylococcal pneumonia, almost 1 in 4 children received antistaphylococcal antibiotic therapy; anti-MRSA therapy was used almost exclusively.
The severity of staphylococcal pneumonia was striking, with >80% of children with S. aureus detected being admitted to intensive care, about 65% requiring invasive mechanical ventilation, and >75% with parapneumonic effusion. These findings are similar to those of prior retrospective studies.4,10 The association between staphylococcal pneumonia and adverse outcomes underscores the importance of prompt institution of antimicrobial therapy targeting S. aureus in high-risk patients. This is noteworthy given recent epidemiological data demonstrating increases in MSSA relative to MRSA infections in children,6 and the known superiority of beta-lactam versus vancomycin for MSSA infections, including pneumonia.11
Although detection of staphylococcal infection was rare, almost a quarter of children received antistaphylococcal therapy; nearly all of these children received anti-MRSA therapy. Confirming a bacterial etiology of pneumonia, however, is challenging. Given the severity associated with staphylococcal pneumonia, it is not surprising that use of antistaphylococcal therapy outpaced staphylococcal detections. Antistaphylococcal therapy was especially common in those with severe pneumonia, suggesting that disease severity is an important factor that influences initial antibiotic treatment decisions. Even so, two children with MRSA detected did not initially receive anti-MRSA therapy, highlighting the challenge of balancing judicious antibiotic selection along with ensuring effective treatment. Perhaps more striking is the finding that 16% of children received antistaphylococcal therapy beyond the first two days of hospitalization, presumably after the initial culture results were available. This suggests that clinicians are reluctant to stop antistaphylococcal therapy when the etiology is unknown, although certain features, such as negative cultures, rapid clinical improvement, and lack of risk factors for staphylococcal disease, may provide important clues to support de-escalation of empiric antibiotic therapy. It is also possible that some antibiotics with antistaphylococcal activity were used for alternative indications (eg, clindamycin for penicillin allergy or concern for aspiration pneumonia).
A simple strategy for tailoring antibiotic treatment is maximizing opportunities to identify a causative pathogen. Despite the very low yield of blood cultures in children with pneumonia overall, bacteremia is more common in children with severe pneumonia and those with parapneumonic effusion, especially when cultures are obtained prior to antibiotic use.12,13 Similarly, obtaining pleural fluid is often therapeutic and significantly improves the chances of identifying a bacterial pathogen.14 Moreover, at least one study suggests that S. aureus is much less likely in cases of culture-negative parapneumonic effusions.15 Institutional guidelines, order sets, and antimicrobial stewardship teams are also effective strategies that can facilitate judicious antibiotic use. In particular, stewardship experts can be very useful in assisting clinicians around de-escalation of therapy.16 Use of procalcitonin, a biomarker associated with bacterial infections,17 and prognostic tools to identify risk for adverse outcomes,18 may also inform treatment decisions and are deserving of further study.
Our study must be considered in
Our study demonstrates a very low prevalence of S. aureus detection among children hospitalized with pneumonia and highlights the association between staphylococcal disease and adverse in-hospital outcomes. We also document important discrepancies between disease prevalence and utilization of antistaphylococcal therapy, especially anti-MRSA therapy. Improved approaches are needed to minimize overuse of antistaphylococcal antibiotics while also ensuring adequate therapy for those who need it.
Disclosures
Drs. Zhu, Edwards, Self, Ampofo, Arnold, McCullers, and Williams report grants from the Centers for Disease Control and Prevention during the conduct of the study. Ms. Frush has nothing to disclose. Dr. Jain has nothing to disclose. Dr. Grijalva reports other from Merck, grants and other from Sanofi, other from Pfizer, grants from CDC, grants from AHRQ, grants from NIH, and grants from Campbell Alliance, outside the submitted work. Dr. Self reports grants from CDC, during the conduct of the study; personal fees from Cempra Pharmaceuticals, grants and personal fees from Ferring Pharmaceuticals, personal fees from BioTest AG, personal fees from Abbott Point of Care, personal fees from Gilead Pharmaceuticals, personal fees from Pfizer, grants from Merck, outside the submitted work. Dr. Thomsen has nothing to disclose. Dr. Ampofo reports grants from CDC, during the conduct of the study; other from GlaxoSmithKline, other from Cubist Pharmaceuticals outside the submitted work; and KA collaborate with BioFire Diagnostics, Inc. (formerly Idaho Technology, Inc.) on several NIH grants. Dr. Pavia reports grants from NAID/NIH, grants from NAID/NIH, grants from CDC, personal fees from WebMD, personal fees from Antimicrobial Therapy Inc., outside the submitted work.
Funding
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number K23AI104779 to D.J.W. and Award 1K23AI113150 to I.P.T., the National Institute of General Medical Sciences under Award K23GM110469 to W.H.S., and the Agency for Healthcare Research and Quality under Award R03HS022342 to C.G.G. The EPIC study was supported by the Influenza Division in the National Center for Immunizations and Respiratory Diseases at the Centers for Disease Control and Prevention through cooperative agreements with each study site and was based on a competitive research funding opportunity. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Institute of Allergy and Infectious Diseases, the National Institute of General Medical Sciences, the Agency for Healthcare Research and Quality, or the Centers for Disease Control and Prevention.
Although Staphylococcus aureus pneumonia is common in children with cystic fibrosis and those with healthcare-associated infections (eg, ventilator-associated pneumonia),1,2 S. aureus is an uncommon cause of community-acquired pneumonia in children. In recent years, concerns have arisen about the increasing frequency and severity of staphylococcal pneumonia, largely fueled by the emergence of community-associated methicillin-resistant S. aureus (MRSA).3,4 Thus, therapy with clindamycin or vancomycin, both active against MRSA, has been recommended when S. aureus is suspected.5 Given the lack of rapid and sensitive approaches to the detection of the etiologies of pneumonia, antibiotic selection is most often empirical, contributing to overuse of anti-MRSA antibiotics. In addition, resistance against these antibiotics, especially clindamycin, has been increasing.6,7
A better understanding of the likelihood of staphylococcal pneumonia would help to optimize empirical antibiotic selection, allowing for judicious use of antistaphylococcal antibiotics, while also avoiding poor outcomes due to delays in effective treatment when S. aureus is present.8 Using data from a multicenter, population-based study of pneumonia hospitalizations in children, we sought to describe the prevalence, clinical characteristics, and in-hospital outcomes of staphylococcal pneumonia and the prevalence of antistaphylococcal antibiotic use.
METHODS
The Etiology of Pneumonia in the Community (EPIC) study was a prospective, active, population-based surveillance study of pneumonia hospitalizations among children (age <18 years) conducted between 2010 and 2012 at three children’s hospitals, including two in Tennessee and one in Utah.9 Children hospitalized with clinical evidence of pneumonia and radiographic evidence confirmed by a blinded review by study radiologists were enrolled. Etiologic assessments included blood analysis for bacterial culture, serology for eight respiratory viruses, pneumococcal and group A streptococcal polymerase chain reaction (PCR), and naso/oro-pharyngeal swabs for PCR for 13 respiratory viruses, Mycoplasma pneumoniae, and Chlamydophila pneumoniae. Data from other clinical specimens (pleural fluid, high-quality endotracheal aspirate, or quantified bronchoalveolar lavage fluid) were also recorded. For this study, we included only children with at least one bacterial culture and complete information about antibiotic use. Those with confirmed fungal pneumonia were excluded. Additional details regarding the study population and methods have been published previously.9
Staphylococcal pneumonia was defined based on the detection of S. aureus by culture (any site) or PCR (pleural fluid only), regardless of codetection of other pathogens. Antibiotic susceptibility profiles were used to classify S. aureus isolates as MRSA or methicillin-sensitive S. aureus (MSSA). The remaining children were classified as nonstaphylococcal pneumonia including children with other bacterial pathogens detected (classified as other bacterial pneumonia, excludes atypical bacteria), atypical bacteria, viruses, and no pathogens detected.
Use of anti-MRSA antibiotics (vancomycin, clindamycin, linezolid, doxycycline, and trimethoprim-sulfamethoxazole) and any antistaphylococcal antibiotics (anti-MRSA agents plus oxacillin, nafcillin, and cefazolin) during and after the first two calendar days of admission was identified by medical record review.
Descriptive statistics included number (%) and median (interquartile range, [IQR]) for categorical and continuous variables, respectively. Baseline clinical characteristics and outcomes were compared between children with staphylococcal versus nonstaphylococcal pneumonia, those with staphylococcal versus other bacterial pneumonia, and those with MRSA versus MSSA pneumonia using Wilcoxon rank-sum and Pearson’s chi-square tests where appropriate. To account for multiple comparisons, we used a Bonferroni corrected P value threshold of <.001 to determine statistical significance.
RESULTS
Of the 2,358 children enrolled in the EPIC study hospitalized with radiographically confirmed pneumonia, 2,146 (91.0%) had ≥1 bacterial culture obtained. Two children with Histoplasma capsulatum fungal infection and six children with incomplete antibiotic utilization data were excluded, yielding a final study population of 2,138 children. Among these, blood samples were obtained from 2,134 (>99%) children for culture, pleural fluid from 87 (4%) children, bronchoalveolar lavage fluid from 31 (1%) children, and endotracheal aspirate from 80 (4%) children. Across all culture types, there were 2,332 initial cultures; 2,150 (92%) were collected within the first 24 hours.
Staphylococcal pneumonia was detected in 23 of the 2,138 children (1% [95% CI 0.7, 1.6]; 17 MRSA, 6 MSSA). Of these, 6/23 (26%) had bacteremia, 12/23 (52%) had a positive pleural fluid, and 9/23 (39%) had a positive culture from bronchoalveolar lavage fluid or endotracheal aspirate; 4/23 (17%) children had S. aureus detected from more than one site. Three children (13%) with S. aureus had a viral codetection, including two with influenza.
Compared with children with nonstaphylococcal pneumonia, those with staphylococcal pneumonia were more likely to have a parapneumonic effusion (78% vs 12%, P < .001), but less likely to have cough (78% vs 95%, P < .001). Other baseline characteristics were similar between the two groups. Children with staphylococcal pneumonia had more adverse outcomes than those without (Table), including longer median length of stay (10 vs 3 days, P < .001), more frequent admission to intensive care (83% vs 21%, P < .001), and more frequent invasive mechanical ventilation (65% vs 7%, P < .001). Similar findings were noted when staphylococcal pneumonia was compared with pneumonia caused due to other bacterial pathogens (n = 124). There were no significant differences in baseline characteristics or clinical course between children with MRSA and MSSA pneumonia, although the numbers were small. Overall, S. aureus was detected in 18/267 (7%) children with parapneumonic effusion and 19/462 (4%) children admitted to intensive care. Importantly, there were no confirmed S. aureus cases among children with less severe pneumonia, defined as lacking both parapneumonic effusion and intensive care admission (n = 1,488).
Overall, 519 children (24%) received antistaphylococcal therapy during their hospitalization (512/519, 99% received anti-MRSA therapy), including 22 of the 23 children with S. aureus detected (the only child without antistaphylococcal therapy had S. aureus detected from a high-quality endotracheal tube aspirate only and also had respiratory syncytial virus detected). Clindamycin was most often used (n = 266, 51%), followed by vancomycin (n = 128, 24%), clindamycin plus vancomycin (n = 83, 16%), and others (n = 42, 8%). During the first two days of hospitalization, 479 children (22%) received antistaphylococcal therapy (477 received anti-MRSA therapy). After the first two days, 351 children (16%) received antistaphylococcal therapy (346/351, 99% received anti-MRSA therapy). Use of antistaphylococcal therapy was very common in those admitted to intensive care (182/462, 39%; all but two received anti-MRSA therapy) and in those requiring invasive mechanical ventilation (103/159, 65%). Among those lacking both parapneumonic effusion and intensive care admission (n = 1488), 232 (16%) received antistaphylococcal therapy.
DISCUSSION
In our large, population-based study of >2,000 children hospitalized with community-acquired pneumonia, S. aureus was identified in only 1% of children. Compared with children with other pneumonia etiologies, staphylococcal pneumonia was associated with increased disease severity. Among the small numbers studied, no differences in outcomes were found between children with MRSA and MSSA disease. Despite the low prevalence of staphylococcal pneumonia, almost 1 in 4 children received antistaphylococcal antibiotic therapy; anti-MRSA therapy was used almost exclusively.
The severity of staphylococcal pneumonia was striking, with >80% of children with S. aureus detected being admitted to intensive care, about 65% requiring invasive mechanical ventilation, and >75% with parapneumonic effusion. These findings are similar to those of prior retrospective studies.4,10 The association between staphylococcal pneumonia and adverse outcomes underscores the importance of prompt institution of antimicrobial therapy targeting S. aureus in high-risk patients. This is noteworthy given recent epidemiological data demonstrating increases in MSSA relative to MRSA infections in children,6 and the known superiority of beta-lactam versus vancomycin for MSSA infections, including pneumonia.11
Although detection of staphylococcal infection was rare, almost a quarter of children received antistaphylococcal therapy; nearly all of these children received anti-MRSA therapy. Confirming a bacterial etiology of pneumonia, however, is challenging. Given the severity associated with staphylococcal pneumonia, it is not surprising that use of antistaphylococcal therapy outpaced staphylococcal detections. Antistaphylococcal therapy was especially common in those with severe pneumonia, suggesting that disease severity is an important factor that influences initial antibiotic treatment decisions. Even so, two children with MRSA detected did not initially receive anti-MRSA therapy, highlighting the challenge of balancing judicious antibiotic selection along with ensuring effective treatment. Perhaps more striking is the finding that 16% of children received antistaphylococcal therapy beyond the first two days of hospitalization, presumably after the initial culture results were available. This suggests that clinicians are reluctant to stop antistaphylococcal therapy when the etiology is unknown, although certain features, such as negative cultures, rapid clinical improvement, and lack of risk factors for staphylococcal disease, may provide important clues to support de-escalation of empiric antibiotic therapy. It is also possible that some antibiotics with antistaphylococcal activity were used for alternative indications (eg, clindamycin for penicillin allergy or concern for aspiration pneumonia).
A simple strategy for tailoring antibiotic treatment is maximizing opportunities to identify a causative pathogen. Despite the very low yield of blood cultures in children with pneumonia overall, bacteremia is more common in children with severe pneumonia and those with parapneumonic effusion, especially when cultures are obtained prior to antibiotic use.12,13 Similarly, obtaining pleural fluid is often therapeutic and significantly improves the chances of identifying a bacterial pathogen.14 Moreover, at least one study suggests that S. aureus is much less likely in cases of culture-negative parapneumonic effusions.15 Institutional guidelines, order sets, and antimicrobial stewardship teams are also effective strategies that can facilitate judicious antibiotic use. In particular, stewardship experts can be very useful in assisting clinicians around de-escalation of therapy.16 Use of procalcitonin, a biomarker associated with bacterial infections,17 and prognostic tools to identify risk for adverse outcomes,18 may also inform treatment decisions and are deserving of further study.
Our study must be considered in
Our study demonstrates a very low prevalence of S. aureus detection among children hospitalized with pneumonia and highlights the association between staphylococcal disease and adverse in-hospital outcomes. We also document important discrepancies between disease prevalence and utilization of antistaphylococcal therapy, especially anti-MRSA therapy. Improved approaches are needed to minimize overuse of antistaphylococcal antibiotics while also ensuring adequate therapy for those who need it.
Disclosures
Drs. Zhu, Edwards, Self, Ampofo, Arnold, McCullers, and Williams report grants from the Centers for Disease Control and Prevention during the conduct of the study. Ms. Frush has nothing to disclose. Dr. Jain has nothing to disclose. Dr. Grijalva reports other from Merck, grants and other from Sanofi, other from Pfizer, grants from CDC, grants from AHRQ, grants from NIH, and grants from Campbell Alliance, outside the submitted work. Dr. Self reports grants from CDC, during the conduct of the study; personal fees from Cempra Pharmaceuticals, grants and personal fees from Ferring Pharmaceuticals, personal fees from BioTest AG, personal fees from Abbott Point of Care, personal fees from Gilead Pharmaceuticals, personal fees from Pfizer, grants from Merck, outside the submitted work. Dr. Thomsen has nothing to disclose. Dr. Ampofo reports grants from CDC, during the conduct of the study; other from GlaxoSmithKline, other from Cubist Pharmaceuticals outside the submitted work; and KA collaborate with BioFire Diagnostics, Inc. (formerly Idaho Technology, Inc.) on several NIH grants. Dr. Pavia reports grants from NAID/NIH, grants from NAID/NIH, grants from CDC, personal fees from WebMD, personal fees from Antimicrobial Therapy Inc., outside the submitted work.
Funding
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number K23AI104779 to D.J.W. and Award 1K23AI113150 to I.P.T., the National Institute of General Medical Sciences under Award K23GM110469 to W.H.S., and the Agency for Healthcare Research and Quality under Award R03HS022342 to C.G.G. The EPIC study was supported by the Influenza Division in the National Center for Immunizations and Respiratory Diseases at the Centers for Disease Control and Prevention through cooperative agreements with each study site and was based on a competitive research funding opportunity. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Institute of Allergy and Infectious Diseases, the National Institute of General Medical Sciences, the Agency for Healthcare Research and Quality, or the Centers for Disease Control and Prevention.
1. Akil N, Muhlebach MS. Biology and management of methicillin resistant Staphylococcus aureus in cystic fibrosis. Pediatr Pulmonol. 2018. doi: 10.1002/ppul.24139. PubMed
2. Srinivasan R, Asselin J, Gildengorin G, Wiener-Kronish J, Flori HR. A prospective study of ventilator-associated pneumonia in children. Pediatrics.
2009;123(4):1108-1115. doi: 10.1542/peds.2008-1211. PubMed
3. Gonzalez BE, Martinez-Aguilar G, Hulten KG, et al. Severe Staphylococcal sepsis in adolescents in the era of community-acquired methicillin-resistant Staphylococcus aureus. Pediatrics. 2005;115(3):642-648. doi: 10.1542/peds.2004-2300. PubMed
4. Carrillo-Marquez MA, Hulten KG, Hammerman W, Lamberth L, Mason EO, Kaplan SL. Staphylococcus aureus pneumonia in children in the era of community-acquired methicillin-resistance at Texas Children’s Hospital. Pediatr Infect Dis J. 2011;30(7):545-550. doi: 10.1097/INF.0b013e31821618be. PubMed
5. Bradley JS, Byington CL, Shah SS, et al. The management of community-acquired pneumonia in infants and children older than 3 months of age: clinical practice guidelines by the pediatric infectious diseases society and the infectious diseases society of America. Clin Infect Dis. 2011;53(7):e25-e76. doi: 10.1093/cid/cir531. PubMed
6. Sutter DE, Milburn E, Chukwuma U, Dzialowy N, Maranich AM, Hospenthal DR. Changing susceptibility of Staphylococcus aureus in a US pediatric population. Pediatrics. 2016;137(4):e20153099–e20153099. doi: 10.1542/peds.2015-3099. PubMed
7. Sakoulas G, Moellering RC, Jr. Increasing antibiotic resistance among methicillin-resistant Staphylococcus aureus strains. Clin Infect Dis. 2008;46(Suppl 5):S360-S367. doi: 10.1086/533592. PubMed
8. Rubinstein E, Kollef MH, Nathwani D. Pneumonia caused by methicillin-resistant
Staphylococcus aureus. Clin Infect Dis. 2008;46(Suppl 5):S378-S385. doi: 10.1086/533594. PubMed
9. Jain S, Williams DJ, Arnold SR, et al. Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med. 2015;372(9):835-845. doi: 10.1056/NEJMoa1405870. PubMed
10. Kallen AJ, Reed C, Patton M, Arnold KE, Finelli L, Hageman J. Staphylococcus aureus community-onset pneumonia in patients admitted to children’s hospitals during autumn and winter of 2006-2007. Epidemiol Infect. 2010;138(5):666-672. doi: 10.1017/S095026880999135X. PubMed
11. González C, Rubio M, Romero-Vivas J, González M, Picazo JJ. Bacteremic pneumonia due to Staphylococcus aureus: A comparison of disease caused by methicillin-resistant and methicillin-susceptible organisms. Clin Infect Dis. 1999;29(5):1171-1177. doi: 10.1086/313440. PubMed
12. Myers AL, Hall M, Williams DJ, et al. Prevalence of bacteremia in hospitalized pediatric patients with community-acquired pneumonia. Pediatr Infect Dis J. 2013;32(7):736-740. doi: 10.1097/INF.0b013e318290bf63. PubMed
13. Iroh Tam PY, Bernstein E, Ma X, Ferrieri P. Blood culture in evaluation of pediatric community-acquired pneumonia: A systematic review and meta-analysis. Hosp Pediatr. 2015;5(6):324-336. doi: 10.1542/hpeds.2014-0138. PubMed
14. Byington CL, Spencer LY, Johnson TA, et al. An epidemiological investigation of a sustained high rate of pediatric parapneumonic empyema: risk factors and microbiological associations. Clin Infect Dis. 2002;34(4):434-440. doi: 10.1086/338460. PubMed
15. Blaschke AJ, Heyrend C, Byington CL, et al. Molecular analysis improves pathogen identifi cation and epidemiologic study of pediatric parapneumonic empyema. Pediatr Infect Dis J. 2011;30(4):289-294. doi: 10.1097/INF.0b013e3182002d14. PubMed
16. Banerjee R, Teng CB, Cunningham SA, et al. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identifi cation and susceptibility testing. Clin Infect Dis. 2015;61(7):1071-1080. doi: 10.1093/cid/civ447. PubMed
17. Stockmann C, Ampofo K, Killpack J, et al. Procalcitonin accurately identifies hospitalized children with low risk of bacterial community-acquired pneumonia. J Pediatr Infect Dis Soc. 2018;7(1):46–53. doi: 10.1093/jpids/piw091. PubMed
18. Williams DJ, Zhu Y, Grijalva CG, et al. Predicting severe pneumonia outcomes in children. Pediatrics. 2016;138(4). doi: 10.1542/peds.2016-1019. PubMed
19. Brogan TV, Hall M, Williams DJ, et al. Variability in processes of care and outcomes among children hospitalized with community-acquired pneumonia. Pediatr Infect Dis J. 2012;31(10):1036-1041. doi: 10.1097/INF.0b013e-31825f2b10. PubMed
1. Akil N, Muhlebach MS. Biology and management of methicillin resistant Staphylococcus aureus in cystic fibrosis. Pediatr Pulmonol. 2018. doi: 10.1002/ppul.24139. PubMed
2. Srinivasan R, Asselin J, Gildengorin G, Wiener-Kronish J, Flori HR. A prospective study of ventilator-associated pneumonia in children. Pediatrics.
2009;123(4):1108-1115. doi: 10.1542/peds.2008-1211. PubMed
3. Gonzalez BE, Martinez-Aguilar G, Hulten KG, et al. Severe Staphylococcal sepsis in adolescents in the era of community-acquired methicillin-resistant Staphylococcus aureus. Pediatrics. 2005;115(3):642-648. doi: 10.1542/peds.2004-2300. PubMed
4. Carrillo-Marquez MA, Hulten KG, Hammerman W, Lamberth L, Mason EO, Kaplan SL. Staphylococcus aureus pneumonia in children in the era of community-acquired methicillin-resistance at Texas Children’s Hospital. Pediatr Infect Dis J. 2011;30(7):545-550. doi: 10.1097/INF.0b013e31821618be. PubMed
5. Bradley JS, Byington CL, Shah SS, et al. The management of community-acquired pneumonia in infants and children older than 3 months of age: clinical practice guidelines by the pediatric infectious diseases society and the infectious diseases society of America. Clin Infect Dis. 2011;53(7):e25-e76. doi: 10.1093/cid/cir531. PubMed
6. Sutter DE, Milburn E, Chukwuma U, Dzialowy N, Maranich AM, Hospenthal DR. Changing susceptibility of Staphylococcus aureus in a US pediatric population. Pediatrics. 2016;137(4):e20153099–e20153099. doi: 10.1542/peds.2015-3099. PubMed
7. Sakoulas G, Moellering RC, Jr. Increasing antibiotic resistance among methicillin-resistant Staphylococcus aureus strains. Clin Infect Dis. 2008;46(Suppl 5):S360-S367. doi: 10.1086/533592. PubMed
8. Rubinstein E, Kollef MH, Nathwani D. Pneumonia caused by methicillin-resistant
Staphylococcus aureus. Clin Infect Dis. 2008;46(Suppl 5):S378-S385. doi: 10.1086/533594. PubMed
9. Jain S, Williams DJ, Arnold SR, et al. Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med. 2015;372(9):835-845. doi: 10.1056/NEJMoa1405870. PubMed
10. Kallen AJ, Reed C, Patton M, Arnold KE, Finelli L, Hageman J. Staphylococcus aureus community-onset pneumonia in patients admitted to children’s hospitals during autumn and winter of 2006-2007. Epidemiol Infect. 2010;138(5):666-672. doi: 10.1017/S095026880999135X. PubMed
11. González C, Rubio M, Romero-Vivas J, González M, Picazo JJ. Bacteremic pneumonia due to Staphylococcus aureus: A comparison of disease caused by methicillin-resistant and methicillin-susceptible organisms. Clin Infect Dis. 1999;29(5):1171-1177. doi: 10.1086/313440. PubMed
12. Myers AL, Hall M, Williams DJ, et al. Prevalence of bacteremia in hospitalized pediatric patients with community-acquired pneumonia. Pediatr Infect Dis J. 2013;32(7):736-740. doi: 10.1097/INF.0b013e318290bf63. PubMed
13. Iroh Tam PY, Bernstein E, Ma X, Ferrieri P. Blood culture in evaluation of pediatric community-acquired pneumonia: A systematic review and meta-analysis. Hosp Pediatr. 2015;5(6):324-336. doi: 10.1542/hpeds.2014-0138. PubMed
14. Byington CL, Spencer LY, Johnson TA, et al. An epidemiological investigation of a sustained high rate of pediatric parapneumonic empyema: risk factors and microbiological associations. Clin Infect Dis. 2002;34(4):434-440. doi: 10.1086/338460. PubMed
15. Blaschke AJ, Heyrend C, Byington CL, et al. Molecular analysis improves pathogen identifi cation and epidemiologic study of pediatric parapneumonic empyema. Pediatr Infect Dis J. 2011;30(4):289-294. doi: 10.1097/INF.0b013e3182002d14. PubMed
16. Banerjee R, Teng CB, Cunningham SA, et al. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identifi cation and susceptibility testing. Clin Infect Dis. 2015;61(7):1071-1080. doi: 10.1093/cid/civ447. PubMed
17. Stockmann C, Ampofo K, Killpack J, et al. Procalcitonin accurately identifies hospitalized children with low risk of bacterial community-acquired pneumonia. J Pediatr Infect Dis Soc. 2018;7(1):46–53. doi: 10.1093/jpids/piw091. PubMed
18. Williams DJ, Zhu Y, Grijalva CG, et al. Predicting severe pneumonia outcomes in children. Pediatrics. 2016;138(4). doi: 10.1542/peds.2016-1019. PubMed
19. Brogan TV, Hall M, Williams DJ, et al. Variability in processes of care and outcomes among children hospitalized with community-acquired pneumonia. Pediatr Infect Dis J. 2012;31(10):1036-1041. doi: 10.1097/INF.0b013e-31825f2b10. PubMed
©2018 Society of Hospital Medicine