User login
Insurers to pay record number of rebates to patients
Health insurance companies are getting ready to disburse a record $1.3 billion in medical loss ratio (MLR) rebates, according to an analysis by the Kaiser Family Foundation.
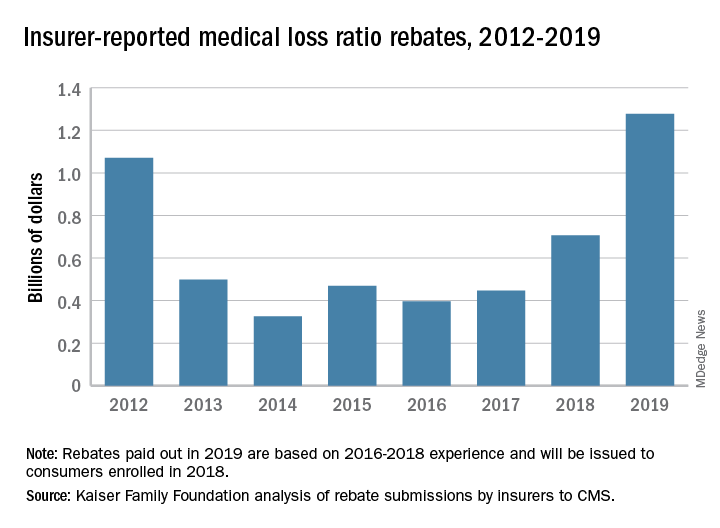
The $1.3 billion surpasses the previous rebate record of $1.1 billion, issued in 2012.
The increase is driven largely by individual market insurers who will pay $743 million in rebates this year, according to the report, which analyzed insurer data submitted to the Centers for Medicare & Medicaid Services. Rebates in the small-group and large-group insurance markets are similar to previous years, with expected paybacks of $250 million from small- and $284 million from large-group markets, according to the Kaiser report. Insurance companies have until September 30, 2019, to start issuing rebates.
The rebates stem from the MLR requirement imposed by the Affordable Care Act (ACA), which limits the amount of premium dollars that can be used for administration, marketing, and profit. Under the health law, companies are required to publicly report how much they spend on health care, quality improvement, and other activities using premium funds. Individual and small-group market insurers must spend at least 80% on health care claims and quality improvement,while large-group plans must spend at least 85%. Rebates are based on a 3-year average of financial data by each insurer.
Patients in the individual insurance market can expect their rebate in either a premium credit or a check. In the large and small group markets, rebates may be split between employee and employer depending on the plan contract.
The volume of rebates differed greatly across the states, with some states paying zero rebates and others paying millions. Virginia insurers for example, will pay the highest number of total rebates ($150 million), followed by Pennsylvania ($130 million) and Florida ($107 million), according to the report. Payments by insurers in the individual market alone ranged from zero dollars in 13 states to $111 million in Virginia. Individual market insurers in Arizona will pay $92 million in rebates to patients, while individual plans in Texas will pay $80 million. Florida insurers will pay the highest in rebates in both the small-group and large-group market at $44 million and $42 million respectively.
The largest rebates within the individual market will come from Centene, HCSC, Cigna, and Highmark. Authors of the report noted that these insurers tend to have higher enrollment and are active in multiple states.
Individual marketplace insurers will likely pay high rebates against next year, based on an individual market that remains strong and profitable, despite the recent elimination of the individual mandate penalty, according to the authors.
Health insurance companies are getting ready to disburse a record $1.3 billion in medical loss ratio (MLR) rebates, according to an analysis by the Kaiser Family Foundation.

The $1.3 billion surpasses the previous rebate record of $1.1 billion, issued in 2012.
The increase is driven largely by individual market insurers who will pay $743 million in rebates this year, according to the report, which analyzed insurer data submitted to the Centers for Medicare & Medicaid Services. Rebates in the small-group and large-group insurance markets are similar to previous years, with expected paybacks of $250 million from small- and $284 million from large-group markets, according to the Kaiser report. Insurance companies have until September 30, 2019, to start issuing rebates.
The rebates stem from the MLR requirement imposed by the Affordable Care Act (ACA), which limits the amount of premium dollars that can be used for administration, marketing, and profit. Under the health law, companies are required to publicly report how much they spend on health care, quality improvement, and other activities using premium funds. Individual and small-group market insurers must spend at least 80% on health care claims and quality improvement,while large-group plans must spend at least 85%. Rebates are based on a 3-year average of financial data by each insurer.
Patients in the individual insurance market can expect their rebate in either a premium credit or a check. In the large and small group markets, rebates may be split between employee and employer depending on the plan contract.
The volume of rebates differed greatly across the states, with some states paying zero rebates and others paying millions. Virginia insurers for example, will pay the highest number of total rebates ($150 million), followed by Pennsylvania ($130 million) and Florida ($107 million), according to the report. Payments by insurers in the individual market alone ranged from zero dollars in 13 states to $111 million in Virginia. Individual market insurers in Arizona will pay $92 million in rebates to patients, while individual plans in Texas will pay $80 million. Florida insurers will pay the highest in rebates in both the small-group and large-group market at $44 million and $42 million respectively.
The largest rebates within the individual market will come from Centene, HCSC, Cigna, and Highmark. Authors of the report noted that these insurers tend to have higher enrollment and are active in multiple states.
Individual marketplace insurers will likely pay high rebates against next year, based on an individual market that remains strong and profitable, despite the recent elimination of the individual mandate penalty, according to the authors.
Health insurance companies are getting ready to disburse a record $1.3 billion in medical loss ratio (MLR) rebates, according to an analysis by the Kaiser Family Foundation.

The $1.3 billion surpasses the previous rebate record of $1.1 billion, issued in 2012.
The increase is driven largely by individual market insurers who will pay $743 million in rebates this year, according to the report, which analyzed insurer data submitted to the Centers for Medicare & Medicaid Services. Rebates in the small-group and large-group insurance markets are similar to previous years, with expected paybacks of $250 million from small- and $284 million from large-group markets, according to the Kaiser report. Insurance companies have until September 30, 2019, to start issuing rebates.
The rebates stem from the MLR requirement imposed by the Affordable Care Act (ACA), which limits the amount of premium dollars that can be used for administration, marketing, and profit. Under the health law, companies are required to publicly report how much they spend on health care, quality improvement, and other activities using premium funds. Individual and small-group market insurers must spend at least 80% on health care claims and quality improvement,while large-group plans must spend at least 85%. Rebates are based on a 3-year average of financial data by each insurer.
Patients in the individual insurance market can expect their rebate in either a premium credit or a check. In the large and small group markets, rebates may be split between employee and employer depending on the plan contract.
The volume of rebates differed greatly across the states, with some states paying zero rebates and others paying millions. Virginia insurers for example, will pay the highest number of total rebates ($150 million), followed by Pennsylvania ($130 million) and Florida ($107 million), according to the report. Payments by insurers in the individual market alone ranged from zero dollars in 13 states to $111 million in Virginia. Individual market insurers in Arizona will pay $92 million in rebates to patients, while individual plans in Texas will pay $80 million. Florida insurers will pay the highest in rebates in both the small-group and large-group market at $44 million and $42 million respectively.
The largest rebates within the individual market will come from Centene, HCSC, Cigna, and Highmark. Authors of the report noted that these insurers tend to have higher enrollment and are active in multiple states.
Individual marketplace insurers will likely pay high rebates against next year, based on an individual market that remains strong and profitable, despite the recent elimination of the individual mandate penalty, according to the authors.
Stem cells gene edited to be HIV resistant treat ALL, but not HIV
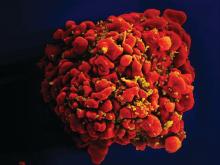
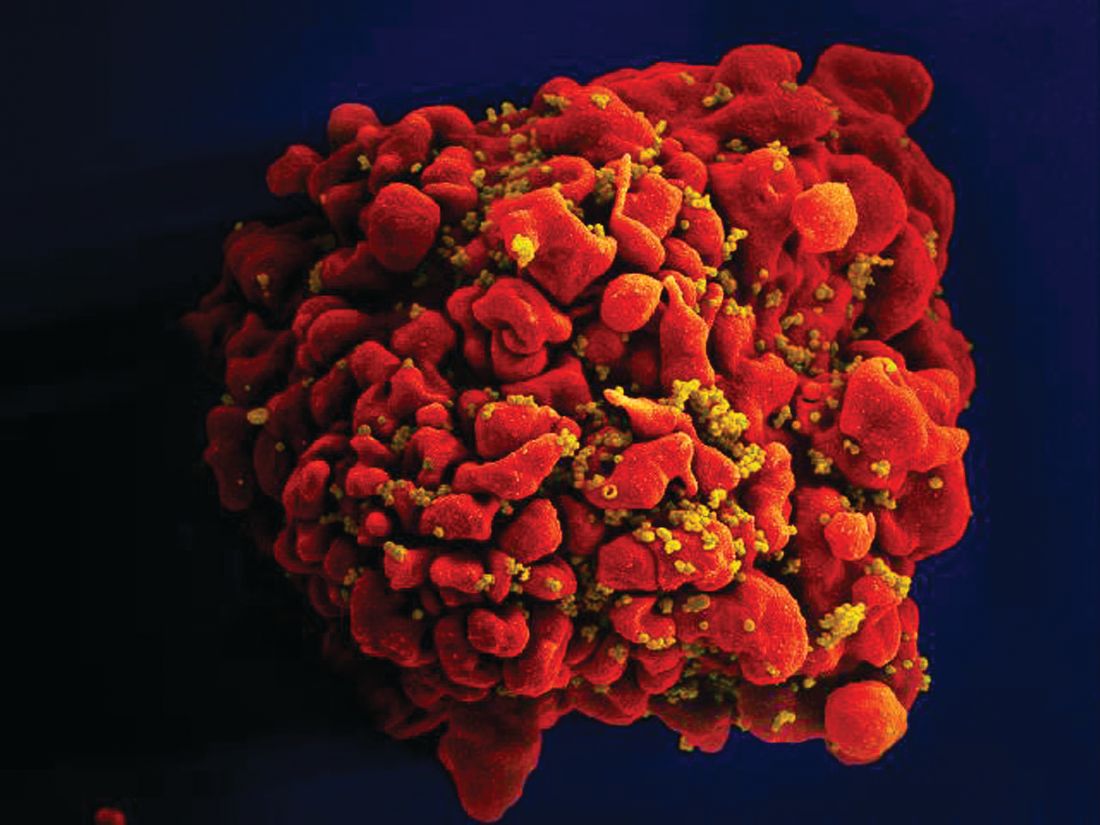
Gene editing of donor stem cells prior to transplantation into a patient with both HIV infection and acute lymphoblastic leukemia (ALL) was safe and effectively treated the patient’s leukemia, but failed to resolve his HIV, investigators reported.
The 27-year-old man received an HLA-matched transplant of hematopoietic stem and progenitor cells (HSPCs) that had been genetically engineered to lack CCR5, a key gateway for HIV entry into cells.
Although the transplant resulted in complete remission of leukemia with full donor chimerism, only about 9% of the posttransplant lymphocytes showed disruption of CCR5, and during a brief trial of antiretroviral therapy interruption his HIV viral load rebounded, reported Hongkui Deng, PhD, and colleagues from Peking University in China.
Although the experiment did not meet its goal of a drug-free HIV remission, it serves as a proof of concept for the use of CRISPR-Cas9 (clustered regularly interspaced palindromic repeats/CRISPR-associated protein 9) gene editing to treat HIV infection, the authors contend.
“These results show the proof of principle that transplantation and long-term engraftment of CRISPR-edited allogeneic HSPCs can be achieved; however, the efficiency of the response was not adequate to achieve the target of cure of HIV-1 infection,” they wrote in a brief report published in the New England Journal of Medicine.
As previously reported, other research groups have investigated genetic editing to mimic a naturally occurring mutation that effectively disables the CCR5 HIV coreceptor, preventing the retrovirus from entering healthy cells. The mutation was first identified in a man named Timothy Brown who came to be known as “the Berlin patient” after he was apparently cured of HIV infection after a bone marrow transplant from a donor who had the mutation.
Dr. Deng and colleagues took advantage of HSPC transplantation, a standard therapy for ALL to see whether it could also have beneficial effects on concomitant HIV infection.
They treated donor HSPCs with CRISPR-Cas9 to ablate CCR5 and then delivered them to the patient along with additional CD34-depleted donor cells from mobilized peripheral blood.
The transplant was a success, with neutrophil engraftment on day 13 and platelet engraftment on day 27, and the leukemia was in morphologic complete remission at week 4 following transplantation. The patient remained in complete remission from leukemia throughout the 19-month follow-up period, with full donor chimerism .
However, when a planned interruption of antiretroviral therapy was carried out at 7 months post transplant, the serum viral load increased to 3 × 107 copies/ml at week 4 following interruption, and the patient was restarted on the drug. His viral levels gradually decreased to undetectable level during the subsequent months.
The investigators noted that 2 weeks after the drug interruption trial was started there was a small increase in the percentage of CCR5 insertion/deletions.
“The low efficiency of gene editing in the patient may be due to the competitive engraftment of the coinfused HSPCs in CD34-depleted cells and the persistence of donor T cells. To further clarify the anti-HIV effect of CCR5-ablated HSPCs, it will be essential to increase the gene-editing efficiency of our CRISPR-Cas9 system and improve the transplantation protocol,” they wrote.
The study was funded by the Beijing Municipal Science and Technology Commission and others (unspecified). All authors reported having nothing to disclose.
SOURCE: Xu L et al. N Engl J Med. 2019. doi: 10.1056/NEJMoa1817426.
Gene editing of donor stem cells prior to transplantation into a patient with both HIV infection and acute lymphoblastic leukemia (ALL) was safe and effectively treated the patient’s leukemia, but failed to resolve his HIV, investigators reported.
The 27-year-old man received an HLA-matched transplant of hematopoietic stem and progenitor cells (HSPCs) that had been genetically engineered to lack CCR5, a key gateway for HIV entry into cells.
Although the transplant resulted in complete remission of leukemia with full donor chimerism, only about 9% of the posttransplant lymphocytes showed disruption of CCR5, and during a brief trial of antiretroviral therapy interruption his HIV viral load rebounded, reported Hongkui Deng, PhD, and colleagues from Peking University in China.
Although the experiment did not meet its goal of a drug-free HIV remission, it serves as a proof of concept for the use of CRISPR-Cas9 (clustered regularly interspaced palindromic repeats/CRISPR-associated protein 9) gene editing to treat HIV infection, the authors contend.
“These results show the proof of principle that transplantation and long-term engraftment of CRISPR-edited allogeneic HSPCs can be achieved; however, the efficiency of the response was not adequate to achieve the target of cure of HIV-1 infection,” they wrote in a brief report published in the New England Journal of Medicine.
As previously reported, other research groups have investigated genetic editing to mimic a naturally occurring mutation that effectively disables the CCR5 HIV coreceptor, preventing the retrovirus from entering healthy cells. The mutation was first identified in a man named Timothy Brown who came to be known as “the Berlin patient” after he was apparently cured of HIV infection after a bone marrow transplant from a donor who had the mutation.
Dr. Deng and colleagues took advantage of HSPC transplantation, a standard therapy for ALL to see whether it could also have beneficial effects on concomitant HIV infection.
They treated donor HSPCs with CRISPR-Cas9 to ablate CCR5 and then delivered them to the patient along with additional CD34-depleted donor cells from mobilized peripheral blood.
The transplant was a success, with neutrophil engraftment on day 13 and platelet engraftment on day 27, and the leukemia was in morphologic complete remission at week 4 following transplantation. The patient remained in complete remission from leukemia throughout the 19-month follow-up period, with full donor chimerism .
However, when a planned interruption of antiretroviral therapy was carried out at 7 months post transplant, the serum viral load increased to 3 × 107 copies/ml at week 4 following interruption, and the patient was restarted on the drug. His viral levels gradually decreased to undetectable level during the subsequent months.
The investigators noted that 2 weeks after the drug interruption trial was started there was a small increase in the percentage of CCR5 insertion/deletions.
“The low efficiency of gene editing in the patient may be due to the competitive engraftment of the coinfused HSPCs in CD34-depleted cells and the persistence of donor T cells. To further clarify the anti-HIV effect of CCR5-ablated HSPCs, it will be essential to increase the gene-editing efficiency of our CRISPR-Cas9 system and improve the transplantation protocol,” they wrote.
The study was funded by the Beijing Municipal Science and Technology Commission and others (unspecified). All authors reported having nothing to disclose.
SOURCE: Xu L et al. N Engl J Med. 2019. doi: 10.1056/NEJMoa1817426.
Gene editing of donor stem cells prior to transplantation into a patient with both HIV infection and acute lymphoblastic leukemia (ALL) was safe and effectively treated the patient’s leukemia, but failed to resolve his HIV, investigators reported.
The 27-year-old man received an HLA-matched transplant of hematopoietic stem and progenitor cells (HSPCs) that had been genetically engineered to lack CCR5, a key gateway for HIV entry into cells.
Although the transplant resulted in complete remission of leukemia with full donor chimerism, only about 9% of the posttransplant lymphocytes showed disruption of CCR5, and during a brief trial of antiretroviral therapy interruption his HIV viral load rebounded, reported Hongkui Deng, PhD, and colleagues from Peking University in China.
Although the experiment did not meet its goal of a drug-free HIV remission, it serves as a proof of concept for the use of CRISPR-Cas9 (clustered regularly interspaced palindromic repeats/CRISPR-associated protein 9) gene editing to treat HIV infection, the authors contend.
“These results show the proof of principle that transplantation and long-term engraftment of CRISPR-edited allogeneic HSPCs can be achieved; however, the efficiency of the response was not adequate to achieve the target of cure of HIV-1 infection,” they wrote in a brief report published in the New England Journal of Medicine.
As previously reported, other research groups have investigated genetic editing to mimic a naturally occurring mutation that effectively disables the CCR5 HIV coreceptor, preventing the retrovirus from entering healthy cells. The mutation was first identified in a man named Timothy Brown who came to be known as “the Berlin patient” after he was apparently cured of HIV infection after a bone marrow transplant from a donor who had the mutation.
Dr. Deng and colleagues took advantage of HSPC transplantation, a standard therapy for ALL to see whether it could also have beneficial effects on concomitant HIV infection.
They treated donor HSPCs with CRISPR-Cas9 to ablate CCR5 and then delivered them to the patient along with additional CD34-depleted donor cells from mobilized peripheral blood.
The transplant was a success, with neutrophil engraftment on day 13 and platelet engraftment on day 27, and the leukemia was in morphologic complete remission at week 4 following transplantation. The patient remained in complete remission from leukemia throughout the 19-month follow-up period, with full donor chimerism .
However, when a planned interruption of antiretroviral therapy was carried out at 7 months post transplant, the serum viral load increased to 3 × 107 copies/ml at week 4 following interruption, and the patient was restarted on the drug. His viral levels gradually decreased to undetectable level during the subsequent months.
The investigators noted that 2 weeks after the drug interruption trial was started there was a small increase in the percentage of CCR5 insertion/deletions.
“The low efficiency of gene editing in the patient may be due to the competitive engraftment of the coinfused HSPCs in CD34-depleted cells and the persistence of donor T cells. To further clarify the anti-HIV effect of CCR5-ablated HSPCs, it will be essential to increase the gene-editing efficiency of our CRISPR-Cas9 system and improve the transplantation protocol,” they wrote.
The study was funded by the Beijing Municipal Science and Technology Commission and others (unspecified). All authors reported having nothing to disclose.
SOURCE: Xu L et al. N Engl J Med. 2019. doi: 10.1056/NEJMoa1817426.
FROM NEW ENGLAND JOURNAL OF MEDICINE
Key clinical point: Donor cells depleted of the HIV coreceptor CCR5 effectively treated ALL, but not HIV.
Major finding: The patient had a sustained complete remission of ALL, but HIV persisted after transplantation.
Study details: Case report of a 27-year-old man with ALL and HIV.
Disclosures: The study was funded by the Beijing Municipal Science and Technology Commission and others (unspecified). All authors reported having nothing to disclose.
Source: Xu L et al. N Engl J Med. 2019. doi: 10.1056/NEJMoa1817426.
No decrease in preterm birth with n-3 fatty acid supplements
according to data published in the New England Journal of Medicine.
Maria Makrides, PhD, of the South Australian Health and Medical Research Institute, North Adelaide, and coauthors wrote there is evidence that n-3 long-chain polyunsaturated fatty acids play an essential role in labor initiation.
“Typical Western diets are relatively low in n-3 long-chain polyunsaturated fatty acids, which leads to a predominance of 2-series prostaglandin substrate in the fetoplacental unit and potentially confers a predisposition to preterm delivery,” they wrote, adding that epidemiologic studies have suggested associations between lower fish consumption in pregnancy and a higher rate of preterm delivery.
In a multicenter, double-blind trial, 5,517 women were randomized to either a daily fish oil supplement containing 900 mg of n-3 long-chain polyunsaturated fatty acids or vegetable oil capsules, from before 20 weeks’ gestation until 34 weeks’ gestation or delivery.
Among the 5,486 pregnancies included in the final analysis, there were no differences between the intervention and control groups in the primary outcome of early preterm delivery, which occurred in 2.2% of pregnancies in the n-3 fatty acid group and 2% of the control group (P = 0.5).
The study also saw no significant differences between the two groups in other outcomes such as the rates of preterm delivery, preterm spontaneous labor, postterm induction, or gestational age at delivery. Similarly, there were no apparent effects of supplementation on maternal and neonatal outcomes including low birth weight, admission to neonatal intensive care, gestational diabetes, postpartum hemorrhage, or preeclampsia.
The analysis did suggest a greater incidence of infants born very large for gestational age – with a birth weight above the 97th percentile – among women in the fatty acid supplement group, but this did not correspond to an increased rate of interventions such as cesarean section or postterm induction.
The authors commented that their finding of more very-large-for-gestational-age babies added to the debate about whether n-3 long-chain polyunsaturated fatty acid supplementation did have a direct impact on fetal growth, although they also noted that it could be a chance finding.
There were also no significant differences between the two groups in serious adverse events, including miscarriage.
The authors noted that the baseline level of n-3 long-chain polyunsaturated fatty acids in the women enrolled in trial may have been higher than in previous studies.
The study was supported by the Australian National Health and Medical Research Council and the Thyne Reid Foundation, with in-kind support from Croda UK and Efamol/Wassen UK. Two authors declared advisory board fees from private industry, and one also declared a patent relating to fatty acids in research. No other conflicts of interest were declared.
SOURCE: Makrides M et al. N Engl J Med. 2019;381:1035-45.
according to data published in the New England Journal of Medicine.
Maria Makrides, PhD, of the South Australian Health and Medical Research Institute, North Adelaide, and coauthors wrote there is evidence that n-3 long-chain polyunsaturated fatty acids play an essential role in labor initiation.
“Typical Western diets are relatively low in n-3 long-chain polyunsaturated fatty acids, which leads to a predominance of 2-series prostaglandin substrate in the fetoplacental unit and potentially confers a predisposition to preterm delivery,” they wrote, adding that epidemiologic studies have suggested associations between lower fish consumption in pregnancy and a higher rate of preterm delivery.
In a multicenter, double-blind trial, 5,517 women were randomized to either a daily fish oil supplement containing 900 mg of n-3 long-chain polyunsaturated fatty acids or vegetable oil capsules, from before 20 weeks’ gestation until 34 weeks’ gestation or delivery.
Among the 5,486 pregnancies included in the final analysis, there were no differences between the intervention and control groups in the primary outcome of early preterm delivery, which occurred in 2.2% of pregnancies in the n-3 fatty acid group and 2% of the control group (P = 0.5).
The study also saw no significant differences between the two groups in other outcomes such as the rates of preterm delivery, preterm spontaneous labor, postterm induction, or gestational age at delivery. Similarly, there were no apparent effects of supplementation on maternal and neonatal outcomes including low birth weight, admission to neonatal intensive care, gestational diabetes, postpartum hemorrhage, or preeclampsia.
The analysis did suggest a greater incidence of infants born very large for gestational age – with a birth weight above the 97th percentile – among women in the fatty acid supplement group, but this did not correspond to an increased rate of interventions such as cesarean section or postterm induction.
The authors commented that their finding of more very-large-for-gestational-age babies added to the debate about whether n-3 long-chain polyunsaturated fatty acid supplementation did have a direct impact on fetal growth, although they also noted that it could be a chance finding.
There were also no significant differences between the two groups in serious adverse events, including miscarriage.
The authors noted that the baseline level of n-3 long-chain polyunsaturated fatty acids in the women enrolled in trial may have been higher than in previous studies.
The study was supported by the Australian National Health and Medical Research Council and the Thyne Reid Foundation, with in-kind support from Croda UK and Efamol/Wassen UK. Two authors declared advisory board fees from private industry, and one also declared a patent relating to fatty acids in research. No other conflicts of interest were declared.
SOURCE: Makrides M et al. N Engl J Med. 2019;381:1035-45.
according to data published in the New England Journal of Medicine.
Maria Makrides, PhD, of the South Australian Health and Medical Research Institute, North Adelaide, and coauthors wrote there is evidence that n-3 long-chain polyunsaturated fatty acids play an essential role in labor initiation.
“Typical Western diets are relatively low in n-3 long-chain polyunsaturated fatty acids, which leads to a predominance of 2-series prostaglandin substrate in the fetoplacental unit and potentially confers a predisposition to preterm delivery,” they wrote, adding that epidemiologic studies have suggested associations between lower fish consumption in pregnancy and a higher rate of preterm delivery.
In a multicenter, double-blind trial, 5,517 women were randomized to either a daily fish oil supplement containing 900 mg of n-3 long-chain polyunsaturated fatty acids or vegetable oil capsules, from before 20 weeks’ gestation until 34 weeks’ gestation or delivery.
Among the 5,486 pregnancies included in the final analysis, there were no differences between the intervention and control groups in the primary outcome of early preterm delivery, which occurred in 2.2% of pregnancies in the n-3 fatty acid group and 2% of the control group (P = 0.5).
The study also saw no significant differences between the two groups in other outcomes such as the rates of preterm delivery, preterm spontaneous labor, postterm induction, or gestational age at delivery. Similarly, there were no apparent effects of supplementation on maternal and neonatal outcomes including low birth weight, admission to neonatal intensive care, gestational diabetes, postpartum hemorrhage, or preeclampsia.
The analysis did suggest a greater incidence of infants born very large for gestational age – with a birth weight above the 97th percentile – among women in the fatty acid supplement group, but this did not correspond to an increased rate of interventions such as cesarean section or postterm induction.
The authors commented that their finding of more very-large-for-gestational-age babies added to the debate about whether n-3 long-chain polyunsaturated fatty acid supplementation did have a direct impact on fetal growth, although they also noted that it could be a chance finding.
There were also no significant differences between the two groups in serious adverse events, including miscarriage.
The authors noted that the baseline level of n-3 long-chain polyunsaturated fatty acids in the women enrolled in trial may have been higher than in previous studies.
The study was supported by the Australian National Health and Medical Research Council and the Thyne Reid Foundation, with in-kind support from Croda UK and Efamol/Wassen UK. Two authors declared advisory board fees from private industry, and one also declared a patent relating to fatty acids in research. No other conflicts of interest were declared.
SOURCE: Makrides M et al. N Engl J Med. 2019;381:1035-45.
FROM THE NEW ENGLAND JOURNAL OF MEDICINE
Key clinical point: No decrease in preterm birth was seen with n-3 fatty acid supplementation during pregnancy, compared with controls.
Major finding: Early preterm delivery occurred in 2.2% of pregnancies in the n-3 fatty acid group and 2% of the control group (P = 0.5).
Study details: A multicenter, double-blind trial in 5,517 women.
Disclosures: The study was supported by the Australian National Health and Medical Research Council and the Thyne Reid Foundation, with in-kind support from Croda UK and Efamol/Wassen UK. Two authors declared advisory board fees from private industry, and one also declared a patent relating to fatty acids in research. No other conflicts of interest were declared.
Source: Makrides M et al. N Engl J Med. 2019;381:1035-45.
iPhone trypophobia and chicken kissin’
Please, no photos
What does the new iPhone have in common with honeycombs and lotus flowers? They all strike terror and nausea in the hearts of trypophobics everywhere.
Trypophobia, in case you haven’t heard, is the fear of irregular patterns of holes or bumps clustered together. It sounds weird, until you look at photos like this and your skin starts to crawl. Now, we can add the iPhone 11 to the list of fear-inducing everyday objects. The new phone design includes three camera lenses, and it’s giving people … issues. Sure, amateur photographers are ecstatic, but social media users collectively shuddered over their keyboards when the tri-camera was revealed.
Trypophobia is not widely studied, but it’s been theorized that the revulsion is a biological instinct against things that look unsafe or diseased. Safe to say this might lead to Apple losing that core demographic – the trypophobe population. They’ll be switching to Androids en masse.
Don’t kiss your chickens after they hatch
All in all, it’s pretty easy to avoid getting salmonella. Refrigerate your food properly. Don’t eat undercooked ground meats. Oh, and don’t kiss the chickens you’ve been raising in your backyard.
Okay, that’s not the only takeaway from a Centers for Disease Control and Prevention update on the 2019 salmonella outbreak that has so far affected just over a thousand people in 49 states. Because the outbreak has been linked to the increased prevalence of backyard poultry, with 67% of patients interviewed reporting contact with chicks and/or ducklings, the CDC has issued a slew of recommendations on how to avoid salmonella.
Some of them are common sense: Don’t let small children handle livestock, and wash your hands after contact. Some are a bit bizarre: Don’t let poultry wander through your house, and don’t eat or drink where livestock roam and live (eww).
Then there’s the gem: Don’t kiss your chickens, or snuggle them and then touch your face and/or mouth.
We know baby chickens or ducks are adorable. And there’s absolutely nothing wrong with loving your livestock like a cat or dog. Just don’t, um, love your livestock.
Dept. of unintended consequences
This week’s case report is brought to you by the entomologists of Texas Medical Center in Houston.
The original problem: Large numbers of birds, such as grackles and pigeons, which may carry diseases and make a mess with their droppings, were gathering in large numbers in Texas Medical Center’s live oak trees. The campus is visited by 10 million people seeking health care each year.
The solution: Cover the trees with nets to prevent the birds from gathering.
The new problem: The lack of predatory birds has “created a haven for a flourishing population of Megalopyge opercularis, commonly referred to as asps,” according to investigators at Rice University. The asp in question happens to be one of North America’s most toxic caterpillars, and they are 7,300% more abundant in the netted trees, compared with nonnetted trees nearby.
The discussion: “I’ve been stung by a lot of things, and an asp sting definitely ranks high up there,” said Mattheau Comerford, one of the investigators. “It feels like a broken bone, and the pain lasts for hours. I was stung on the wrist, and the pain traveled up my arm, into my arm pit, and my jaw started to feel pain.”
The LOTME recommendation: In this case, the rats with wings … er, we mean pigeons, seem to be the lesser of two evils. Of course, compared with poisonous caterpillars, even kissing a chicken would be the lesser of two evils.

Please, no photos
What does the new iPhone have in common with honeycombs and lotus flowers? They all strike terror and nausea in the hearts of trypophobics everywhere.
Trypophobia, in case you haven’t heard, is the fear of irregular patterns of holes or bumps clustered together. It sounds weird, until you look at photos like this and your skin starts to crawl. Now, we can add the iPhone 11 to the list of fear-inducing everyday objects. The new phone design includes three camera lenses, and it’s giving people … issues. Sure, amateur photographers are ecstatic, but social media users collectively shuddered over their keyboards when the tri-camera was revealed.
Trypophobia is not widely studied, but it’s been theorized that the revulsion is a biological instinct against things that look unsafe or diseased. Safe to say this might lead to Apple losing that core demographic – the trypophobe population. They’ll be switching to Androids en masse.
Don’t kiss your chickens after they hatch
All in all, it’s pretty easy to avoid getting salmonella. Refrigerate your food properly. Don’t eat undercooked ground meats. Oh, and don’t kiss the chickens you’ve been raising in your backyard.
Okay, that’s not the only takeaway from a Centers for Disease Control and Prevention update on the 2019 salmonella outbreak that has so far affected just over a thousand people in 49 states. Because the outbreak has been linked to the increased prevalence of backyard poultry, with 67% of patients interviewed reporting contact with chicks and/or ducklings, the CDC has issued a slew of recommendations on how to avoid salmonella.
Some of them are common sense: Don’t let small children handle livestock, and wash your hands after contact. Some are a bit bizarre: Don’t let poultry wander through your house, and don’t eat or drink where livestock roam and live (eww).
Then there’s the gem: Don’t kiss your chickens, or snuggle them and then touch your face and/or mouth.
We know baby chickens or ducks are adorable. And there’s absolutely nothing wrong with loving your livestock like a cat or dog. Just don’t, um, love your livestock.
Dept. of unintended consequences
This week’s case report is brought to you by the entomologists of Texas Medical Center in Houston.
The original problem: Large numbers of birds, such as grackles and pigeons, which may carry diseases and make a mess with their droppings, were gathering in large numbers in Texas Medical Center’s live oak trees. The campus is visited by 10 million people seeking health care each year.
The solution: Cover the trees with nets to prevent the birds from gathering.
The new problem: The lack of predatory birds has “created a haven for a flourishing population of Megalopyge opercularis, commonly referred to as asps,” according to investigators at Rice University. The asp in question happens to be one of North America’s most toxic caterpillars, and they are 7,300% more abundant in the netted trees, compared with nonnetted trees nearby.
The discussion: “I’ve been stung by a lot of things, and an asp sting definitely ranks high up there,” said Mattheau Comerford, one of the investigators. “It feels like a broken bone, and the pain lasts for hours. I was stung on the wrist, and the pain traveled up my arm, into my arm pit, and my jaw started to feel pain.”
The LOTME recommendation: In this case, the rats with wings … er, we mean pigeons, seem to be the lesser of two evils. Of course, compared with poisonous caterpillars, even kissing a chicken would be the lesser of two evils.

Please, no photos
What does the new iPhone have in common with honeycombs and lotus flowers? They all strike terror and nausea in the hearts of trypophobics everywhere.
Trypophobia, in case you haven’t heard, is the fear of irregular patterns of holes or bumps clustered together. It sounds weird, until you look at photos like this and your skin starts to crawl. Now, we can add the iPhone 11 to the list of fear-inducing everyday objects. The new phone design includes three camera lenses, and it’s giving people … issues. Sure, amateur photographers are ecstatic, but social media users collectively shuddered over their keyboards when the tri-camera was revealed.
Trypophobia is not widely studied, but it’s been theorized that the revulsion is a biological instinct against things that look unsafe or diseased. Safe to say this might lead to Apple losing that core demographic – the trypophobe population. They’ll be switching to Androids en masse.
Don’t kiss your chickens after they hatch
All in all, it’s pretty easy to avoid getting salmonella. Refrigerate your food properly. Don’t eat undercooked ground meats. Oh, and don’t kiss the chickens you’ve been raising in your backyard.
Okay, that’s not the only takeaway from a Centers for Disease Control and Prevention update on the 2019 salmonella outbreak that has so far affected just over a thousand people in 49 states. Because the outbreak has been linked to the increased prevalence of backyard poultry, with 67% of patients interviewed reporting contact with chicks and/or ducklings, the CDC has issued a slew of recommendations on how to avoid salmonella.
Some of them are common sense: Don’t let small children handle livestock, and wash your hands after contact. Some are a bit bizarre: Don’t let poultry wander through your house, and don’t eat or drink where livestock roam and live (eww).
Then there’s the gem: Don’t kiss your chickens, or snuggle them and then touch your face and/or mouth.
We know baby chickens or ducks are adorable. And there’s absolutely nothing wrong with loving your livestock like a cat or dog. Just don’t, um, love your livestock.
Dept. of unintended consequences
This week’s case report is brought to you by the entomologists of Texas Medical Center in Houston.
The original problem: Large numbers of birds, such as grackles and pigeons, which may carry diseases and make a mess with their droppings, were gathering in large numbers in Texas Medical Center’s live oak trees. The campus is visited by 10 million people seeking health care each year.
The solution: Cover the trees with nets to prevent the birds from gathering.
The new problem: The lack of predatory birds has “created a haven for a flourishing population of Megalopyge opercularis, commonly referred to as asps,” according to investigators at Rice University. The asp in question happens to be one of North America’s most toxic caterpillars, and they are 7,300% more abundant in the netted trees, compared with nonnetted trees nearby.
The discussion: “I’ve been stung by a lot of things, and an asp sting definitely ranks high up there,” said Mattheau Comerford, one of the investigators. “It feels like a broken bone, and the pain lasts for hours. I was stung on the wrist, and the pain traveled up my arm, into my arm pit, and my jaw started to feel pain.”
The LOTME recommendation: In this case, the rats with wings … er, we mean pigeons, seem to be the lesser of two evils. Of course, compared with poisonous caterpillars, even kissing a chicken would be the lesser of two evils.

Review your insurance
Insurance, so goes the hoary cliché, is the one product you buy hoping never to use. While no one enjoys foreseeing unforeseeable calamities, if you haven’t reviewed your insurance coverage recently, there is no time like the present.
, but the cost has become prohibitive in many areas, when insurers are willing to write them at all. “Claims made” policies are cheaper and provide the same protection, but only while coverage is in effect. You will need “tail” coverage against belated claims after your policy lapses, but many companies provide free tail coverage if you are retiring. If you are simply switching workplaces (or policies), ask your new insurer about “nose” coverage, for claims involving acts that occurred before the new policy takes effect.
Other alternatives are gaining popularity as the demand for reasonably priced insurance increases. The most common, known as reciprocal exchanges, are very similar to traditional insurers, but require policyholders to make capital contributions in addition to payment of premiums, at least in their early stages. You get your investment back, with interest, when (if) the exchange becomes solvent.
Another option, called a captive, is a company formed by a consortium of medical practices to write their own insurance policies. All participants are shareholders, and all premiums (less administrative expenses) go toward building the security of the captive. Most captives purchase reinsurance to protect against catastrophic losses. If all goes well, individual owners sell their shares at retirement for a profit, which has grown tax-free in the interim.
Those willing to shoulder more risk might consider a risk retention group (RRG), a sort of combination of an exchange and a captive. Again, the owners are the insureds themselves, but all responsibility for management and adequate funding falls on their shoulders, and reinsurance is not usually an option. Most medical malpractice RRGs are licensed in Vermont or South Carolina, because of favorable laws in those states, but can be based in any state that allows them (36 at this writing). RRGs provide profit opportunities not available with traditional insurance, but there is risk: A few large claims could eat up all the profits, or even put owners in a financial hole.
Malpractice insurance requirements will remain fairly static throughout your career, but other insurance needs evolve over time. A good example is life insurance: As retirement savings increase, the need for life insurance decreases – especially expensive “whole life” coverage, which can often be eliminated or converted to cheaper “term” insurance.
Health insurance premiums continue to soar, but the Affordable Care Act might offer a favorable alternative for your office policy. If you are considering that, the Centers for Medicare & Medicaid Services maintains a website summarizing the various options for employers.
Worker compensation insurance is mandatory in most states and heavily regulated, so there is little wiggle room. However, some states do not require you, as the employer, to cover yourself, so eliminating that coverage could save you a substantial amount. This is only worth considering, of course, if you’re in excellent health and have very good personal health and disability coverage.
Disability insurance is not something to skimp on, but if you are approaching retirement age and have no major health issues, you may be able to decrease your coverage, or even eliminate it entirely if your retirement plan is far enough along.
Liability insurance is likewise no place to pinch pennies, but you might be able to add an “umbrella” policy providing comprehensive catastrophic coverage, which may allow you to decrease your regular coverage, or raise your deductible limits.
Two additional policies to consider are office overhead insurance, to cover the costs of keeping your office open should you be temporarily incapacitated, and employee practices liability insurance (EPLI), which protects you from lawsuits brought by militant or disgruntled employees. I covered EPLI in detail several months ago.
If you are over 50, I strongly recommend long-term-care insurance as well. It’s relatively inexpensive if you buy it while you’re still healthy, and it could save you and your heirs a load of money and aggravation on the other end. If you have shouldered the expense of caring for a chronically ill parent or grandparent, you know what I’m talking about. More about that next month.
Dr. Eastern practices dermatology and dermatologic surgery in Belleville, N.J. He is the author of numerous articles and textbook chapters, and is a longtime monthly columnist for Dermatology News. Write to him at dermnews@mdedge.com.
Insurance, so goes the hoary cliché, is the one product you buy hoping never to use. While no one enjoys foreseeing unforeseeable calamities, if you haven’t reviewed your insurance coverage recently, there is no time like the present.
, but the cost has become prohibitive in many areas, when insurers are willing to write them at all. “Claims made” policies are cheaper and provide the same protection, but only while coverage is in effect. You will need “tail” coverage against belated claims after your policy lapses, but many companies provide free tail coverage if you are retiring. If you are simply switching workplaces (or policies), ask your new insurer about “nose” coverage, for claims involving acts that occurred before the new policy takes effect.
Other alternatives are gaining popularity as the demand for reasonably priced insurance increases. The most common, known as reciprocal exchanges, are very similar to traditional insurers, but require policyholders to make capital contributions in addition to payment of premiums, at least in their early stages. You get your investment back, with interest, when (if) the exchange becomes solvent.
Another option, called a captive, is a company formed by a consortium of medical practices to write their own insurance policies. All participants are shareholders, and all premiums (less administrative expenses) go toward building the security of the captive. Most captives purchase reinsurance to protect against catastrophic losses. If all goes well, individual owners sell their shares at retirement for a profit, which has grown tax-free in the interim.
Those willing to shoulder more risk might consider a risk retention group (RRG), a sort of combination of an exchange and a captive. Again, the owners are the insureds themselves, but all responsibility for management and adequate funding falls on their shoulders, and reinsurance is not usually an option. Most medical malpractice RRGs are licensed in Vermont or South Carolina, because of favorable laws in those states, but can be based in any state that allows them (36 at this writing). RRGs provide profit opportunities not available with traditional insurance, but there is risk: A few large claims could eat up all the profits, or even put owners in a financial hole.
Malpractice insurance requirements will remain fairly static throughout your career, but other insurance needs evolve over time. A good example is life insurance: As retirement savings increase, the need for life insurance decreases – especially expensive “whole life” coverage, which can often be eliminated or converted to cheaper “term” insurance.
Health insurance premiums continue to soar, but the Affordable Care Act might offer a favorable alternative for your office policy. If you are considering that, the Centers for Medicare & Medicaid Services maintains a website summarizing the various options for employers.
Worker compensation insurance is mandatory in most states and heavily regulated, so there is little wiggle room. However, some states do not require you, as the employer, to cover yourself, so eliminating that coverage could save you a substantial amount. This is only worth considering, of course, if you’re in excellent health and have very good personal health and disability coverage.
Disability insurance is not something to skimp on, but if you are approaching retirement age and have no major health issues, you may be able to decrease your coverage, or even eliminate it entirely if your retirement plan is far enough along.
Liability insurance is likewise no place to pinch pennies, but you might be able to add an “umbrella” policy providing comprehensive catastrophic coverage, which may allow you to decrease your regular coverage, or raise your deductible limits.
Two additional policies to consider are office overhead insurance, to cover the costs of keeping your office open should you be temporarily incapacitated, and employee practices liability insurance (EPLI), which protects you from lawsuits brought by militant or disgruntled employees. I covered EPLI in detail several months ago.
If you are over 50, I strongly recommend long-term-care insurance as well. It’s relatively inexpensive if you buy it while you’re still healthy, and it could save you and your heirs a load of money and aggravation on the other end. If you have shouldered the expense of caring for a chronically ill parent or grandparent, you know what I’m talking about. More about that next month.
Dr. Eastern practices dermatology and dermatologic surgery in Belleville, N.J. He is the author of numerous articles and textbook chapters, and is a longtime monthly columnist for Dermatology News. Write to him at dermnews@mdedge.com.
Insurance, so goes the hoary cliché, is the one product you buy hoping never to use. While no one enjoys foreseeing unforeseeable calamities, if you haven’t reviewed your insurance coverage recently, there is no time like the present.
, but the cost has become prohibitive in many areas, when insurers are willing to write them at all. “Claims made” policies are cheaper and provide the same protection, but only while coverage is in effect. You will need “tail” coverage against belated claims after your policy lapses, but many companies provide free tail coverage if you are retiring. If you are simply switching workplaces (or policies), ask your new insurer about “nose” coverage, for claims involving acts that occurred before the new policy takes effect.
Other alternatives are gaining popularity as the demand for reasonably priced insurance increases. The most common, known as reciprocal exchanges, are very similar to traditional insurers, but require policyholders to make capital contributions in addition to payment of premiums, at least in their early stages. You get your investment back, with interest, when (if) the exchange becomes solvent.
Another option, called a captive, is a company formed by a consortium of medical practices to write their own insurance policies. All participants are shareholders, and all premiums (less administrative expenses) go toward building the security of the captive. Most captives purchase reinsurance to protect against catastrophic losses. If all goes well, individual owners sell their shares at retirement for a profit, which has grown tax-free in the interim.
Those willing to shoulder more risk might consider a risk retention group (RRG), a sort of combination of an exchange and a captive. Again, the owners are the insureds themselves, but all responsibility for management and adequate funding falls on their shoulders, and reinsurance is not usually an option. Most medical malpractice RRGs are licensed in Vermont or South Carolina, because of favorable laws in those states, but can be based in any state that allows them (36 at this writing). RRGs provide profit opportunities not available with traditional insurance, but there is risk: A few large claims could eat up all the profits, or even put owners in a financial hole.
Malpractice insurance requirements will remain fairly static throughout your career, but other insurance needs evolve over time. A good example is life insurance: As retirement savings increase, the need for life insurance decreases – especially expensive “whole life” coverage, which can often be eliminated or converted to cheaper “term” insurance.
Health insurance premiums continue to soar, but the Affordable Care Act might offer a favorable alternative for your office policy. If you are considering that, the Centers for Medicare & Medicaid Services maintains a website summarizing the various options for employers.
Worker compensation insurance is mandatory in most states and heavily regulated, so there is little wiggle room. However, some states do not require you, as the employer, to cover yourself, so eliminating that coverage could save you a substantial amount. This is only worth considering, of course, if you’re in excellent health and have very good personal health and disability coverage.
Disability insurance is not something to skimp on, but if you are approaching retirement age and have no major health issues, you may be able to decrease your coverage, or even eliminate it entirely if your retirement plan is far enough along.
Liability insurance is likewise no place to pinch pennies, but you might be able to add an “umbrella” policy providing comprehensive catastrophic coverage, which may allow you to decrease your regular coverage, or raise your deductible limits.
Two additional policies to consider are office overhead insurance, to cover the costs of keeping your office open should you be temporarily incapacitated, and employee practices liability insurance (EPLI), which protects you from lawsuits brought by militant or disgruntled employees. I covered EPLI in detail several months ago.
If you are over 50, I strongly recommend long-term-care insurance as well. It’s relatively inexpensive if you buy it while you’re still healthy, and it could save you and your heirs a load of money and aggravation on the other end. If you have shouldered the expense of caring for a chronically ill parent or grandparent, you know what I’m talking about. More about that next month.
Dr. Eastern practices dermatology and dermatologic surgery in Belleville, N.J. He is the author of numerous articles and textbook chapters, and is a longtime monthly columnist for Dermatology News. Write to him at dermnews@mdedge.com.
Colorectal screening cost effective in cystic fibrosis patients
Screening for colorectal cancer in patients with cystic fibrosis is cost effective, and should be started at a younger age and performed more often, new research suggests.
While colorectal cancer (CRC) screening traditionally begins at age 50 years in people at average risk for the disease, those at high risk usually begin undergoing colonoscopies at an earlier age. Patients with cystic fibrosis fall under the latter category, wrote Andrea Gini, of the department of public health at Erasmus Medical Center in Rotterdam, the Netherlands, and colleagues, with an incidence of CRC up to 30 times higher than the general population, but their shorter lifespan has led to a “different trade-off between the benefits and harms of CRC screening.”
Between 2000 and 2015, the median predicted survival age for patients with cystic fibrosis increased from 33.3 years to 41.7 years; this increased survival has brought increased risk for other diseases, particularly in the GI tract, Mr. Gini and colleagues wrote in Gastroenterology. By using the Microsimulation Screening Analysis–Colon model – a joint project between Erasmus Medical Center and Memorial Sloan Kettering Cancer Center in New York – the investigators assessed the cost-effectiveness of CRC screening in patients with cystic fibrosis.
Three cohorts of 10 million patients each were simulated, with one cohort having undergone transplant, one cohort not having transplant, and one cohort of individuals without cystic fibrosis. The simulated patient age was 30 years in 2017. A total of 76 different colonoscopy-screening strategies were assessed, with each differing in screening interval (3, 5, or 10 years for colonoscopy), age to start screening (30, 35, 40, 45, or 50 years), and age to end screening (55, 60, 65, 70, or 75 years). The optimal screening strategy was determined based on a willingness-to-pay threshold of $100,000 per life-year gained, the investigators wrote.
In the absence of screening, the mortality rate for nontransplant cystic fibrosis patients was 19.1 per 1,000 people, and the rate for cystic fibrosis patients who had undergone transplant was 22.3 per 1,000 people. The standard screening strategy prevented more than 73% of CRC deaths in the general population, 66% of deaths in nontransplant cystic fibrosis patients, and 36% of deaths in cystic fibrosis patients with transplant; however, the model predicted that only 22% of individuals who received a transplant and 36% of those who did not would reach the age of 50 years.
According to the model, the optimal colonoscopy-screening strategy for nontransplant patients was one screen every 5 years, starting at 40 and screening until the age of 75. The incremental cost-effectiveness ratio (ICER) was $84,000 per life-year gained; CRC incidence was reduced by 52% and CRC mortality was reduced by 79%. For transplant patients, the best strategy was one screen every 3 years between the ages of 35 and 55, which reduced CRC mortality by 82% at an ICER of $71,000 per life-year gained.
In a separate analysis of fecal immunochemical testing, a less-demanding alternative to colonoscopy, the optimal screening strategy was an annual test between the age of 35 and 75 years for nontransplant cystic fibrosis patients, for an ICER of $47,000 per life-year gained and a CRC mortality reduction of 78%. The best strategy for transplant patients was once a year between the ages of 30 and 60, which reduced CRC mortality by 77% at an ICER of $86,000 per life-year gained. While fecal immunochemical testing may be more cost effective than colonoscopy, “specific evidence of its performance in the cystic fibrosis population is required before considering this screening modality,” the investigators noted.
“This study indicates that there is benefit to earlier CRC screening in the cystic fibrosis population and [that it] can be done at acceptable costs,” the investigators wrote. “The findings of this analysis support clinicians, researchers, and policy makers who aim to define a tailored CRC screening for individuals with cystic fibrosis in the United States.”
The study was funded by the Cystic Fibrosis Foundation, the Cancer Intervention and Surveillance Modeling Network consortium, and Memorial Sloan Kettering Cancer Center. The investigators reported no conflicts of interest.
Help your patients understand what do expect during and how to prepare for a colonoscopy by sharing AGA’s patient education at https://www.gastro.org/practice-guidance/gi-patient-center/topic/colonoscopy.
SOURCE: Gini A et al. Gastroenterology. 2017 Dec 27. doi: 10.1053/j.gastro.2017.12.011.
Screening for colorectal cancer in patients with cystic fibrosis is cost effective, and should be started at a younger age and performed more often, new research suggests.
While colorectal cancer (CRC) screening traditionally begins at age 50 years in people at average risk for the disease, those at high risk usually begin undergoing colonoscopies at an earlier age. Patients with cystic fibrosis fall under the latter category, wrote Andrea Gini, of the department of public health at Erasmus Medical Center in Rotterdam, the Netherlands, and colleagues, with an incidence of CRC up to 30 times higher than the general population, but their shorter lifespan has led to a “different trade-off between the benefits and harms of CRC screening.”
Between 2000 and 2015, the median predicted survival age for patients with cystic fibrosis increased from 33.3 years to 41.7 years; this increased survival has brought increased risk for other diseases, particularly in the GI tract, Mr. Gini and colleagues wrote in Gastroenterology. By using the Microsimulation Screening Analysis–Colon model – a joint project between Erasmus Medical Center and Memorial Sloan Kettering Cancer Center in New York – the investigators assessed the cost-effectiveness of CRC screening in patients with cystic fibrosis.
Three cohorts of 10 million patients each were simulated, with one cohort having undergone transplant, one cohort not having transplant, and one cohort of individuals without cystic fibrosis. The simulated patient age was 30 years in 2017. A total of 76 different colonoscopy-screening strategies were assessed, with each differing in screening interval (3, 5, or 10 years for colonoscopy), age to start screening (30, 35, 40, 45, or 50 years), and age to end screening (55, 60, 65, 70, or 75 years). The optimal screening strategy was determined based on a willingness-to-pay threshold of $100,000 per life-year gained, the investigators wrote.
In the absence of screening, the mortality rate for nontransplant cystic fibrosis patients was 19.1 per 1,000 people, and the rate for cystic fibrosis patients who had undergone transplant was 22.3 per 1,000 people. The standard screening strategy prevented more than 73% of CRC deaths in the general population, 66% of deaths in nontransplant cystic fibrosis patients, and 36% of deaths in cystic fibrosis patients with transplant; however, the model predicted that only 22% of individuals who received a transplant and 36% of those who did not would reach the age of 50 years.
According to the model, the optimal colonoscopy-screening strategy for nontransplant patients was one screen every 5 years, starting at 40 and screening until the age of 75. The incremental cost-effectiveness ratio (ICER) was $84,000 per life-year gained; CRC incidence was reduced by 52% and CRC mortality was reduced by 79%. For transplant patients, the best strategy was one screen every 3 years between the ages of 35 and 55, which reduced CRC mortality by 82% at an ICER of $71,000 per life-year gained.
In a separate analysis of fecal immunochemical testing, a less-demanding alternative to colonoscopy, the optimal screening strategy was an annual test between the age of 35 and 75 years for nontransplant cystic fibrosis patients, for an ICER of $47,000 per life-year gained and a CRC mortality reduction of 78%. The best strategy for transplant patients was once a year between the ages of 30 and 60, which reduced CRC mortality by 77% at an ICER of $86,000 per life-year gained. While fecal immunochemical testing may be more cost effective than colonoscopy, “specific evidence of its performance in the cystic fibrosis population is required before considering this screening modality,” the investigators noted.
“This study indicates that there is benefit to earlier CRC screening in the cystic fibrosis population and [that it] can be done at acceptable costs,” the investigators wrote. “The findings of this analysis support clinicians, researchers, and policy makers who aim to define a tailored CRC screening for individuals with cystic fibrosis in the United States.”
The study was funded by the Cystic Fibrosis Foundation, the Cancer Intervention and Surveillance Modeling Network consortium, and Memorial Sloan Kettering Cancer Center. The investigators reported no conflicts of interest.
Help your patients understand what do expect during and how to prepare for a colonoscopy by sharing AGA’s patient education at https://www.gastro.org/practice-guidance/gi-patient-center/topic/colonoscopy.
SOURCE: Gini A et al. Gastroenterology. 2017 Dec 27. doi: 10.1053/j.gastro.2017.12.011.
Screening for colorectal cancer in patients with cystic fibrosis is cost effective, and should be started at a younger age and performed more often, new research suggests.
While colorectal cancer (CRC) screening traditionally begins at age 50 years in people at average risk for the disease, those at high risk usually begin undergoing colonoscopies at an earlier age. Patients with cystic fibrosis fall under the latter category, wrote Andrea Gini, of the department of public health at Erasmus Medical Center in Rotterdam, the Netherlands, and colleagues, with an incidence of CRC up to 30 times higher than the general population, but their shorter lifespan has led to a “different trade-off between the benefits and harms of CRC screening.”
Between 2000 and 2015, the median predicted survival age for patients with cystic fibrosis increased from 33.3 years to 41.7 years; this increased survival has brought increased risk for other diseases, particularly in the GI tract, Mr. Gini and colleagues wrote in Gastroenterology. By using the Microsimulation Screening Analysis–Colon model – a joint project between Erasmus Medical Center and Memorial Sloan Kettering Cancer Center in New York – the investigators assessed the cost-effectiveness of CRC screening in patients with cystic fibrosis.
Three cohorts of 10 million patients each were simulated, with one cohort having undergone transplant, one cohort not having transplant, and one cohort of individuals without cystic fibrosis. The simulated patient age was 30 years in 2017. A total of 76 different colonoscopy-screening strategies were assessed, with each differing in screening interval (3, 5, or 10 years for colonoscopy), age to start screening (30, 35, 40, 45, or 50 years), and age to end screening (55, 60, 65, 70, or 75 years). The optimal screening strategy was determined based on a willingness-to-pay threshold of $100,000 per life-year gained, the investigators wrote.
In the absence of screening, the mortality rate for nontransplant cystic fibrosis patients was 19.1 per 1,000 people, and the rate for cystic fibrosis patients who had undergone transplant was 22.3 per 1,000 people. The standard screening strategy prevented more than 73% of CRC deaths in the general population, 66% of deaths in nontransplant cystic fibrosis patients, and 36% of deaths in cystic fibrosis patients with transplant; however, the model predicted that only 22% of individuals who received a transplant and 36% of those who did not would reach the age of 50 years.
According to the model, the optimal colonoscopy-screening strategy for nontransplant patients was one screen every 5 years, starting at 40 and screening until the age of 75. The incremental cost-effectiveness ratio (ICER) was $84,000 per life-year gained; CRC incidence was reduced by 52% and CRC mortality was reduced by 79%. For transplant patients, the best strategy was one screen every 3 years between the ages of 35 and 55, which reduced CRC mortality by 82% at an ICER of $71,000 per life-year gained.
In a separate analysis of fecal immunochemical testing, a less-demanding alternative to colonoscopy, the optimal screening strategy was an annual test between the age of 35 and 75 years for nontransplant cystic fibrosis patients, for an ICER of $47,000 per life-year gained and a CRC mortality reduction of 78%. The best strategy for transplant patients was once a year between the ages of 30 and 60, which reduced CRC mortality by 77% at an ICER of $86,000 per life-year gained. While fecal immunochemical testing may be more cost effective than colonoscopy, “specific evidence of its performance in the cystic fibrosis population is required before considering this screening modality,” the investigators noted.
“This study indicates that there is benefit to earlier CRC screening in the cystic fibrosis population and [that it] can be done at acceptable costs,” the investigators wrote. “The findings of this analysis support clinicians, researchers, and policy makers who aim to define a tailored CRC screening for individuals with cystic fibrosis in the United States.”
The study was funded by the Cystic Fibrosis Foundation, the Cancer Intervention and Surveillance Modeling Network consortium, and Memorial Sloan Kettering Cancer Center. The investigators reported no conflicts of interest.
Help your patients understand what do expect during and how to prepare for a colonoscopy by sharing AGA’s patient education at https://www.gastro.org/practice-guidance/gi-patient-center/topic/colonoscopy.
SOURCE: Gini A et al. Gastroenterology. 2017 Dec 27. doi: 10.1053/j.gastro.2017.12.011.
FROM GASTROENTEROLOGY
Many experimental drugs veer off course when targeting cancer
Clinical trials of novel cancer drugs miss their marks far more often than they hit them, in part because the drugs themselves may be aimed at the wrong targets or the targets themselves may not be that important in the first place, investigators have found.
Using CRISPR (clustered regularly interspaced palindromic repeats) gene editing to study the effects of 10 drugs that are in development targeting six proteins ostensibly crucial to the health or survival of cancer cells, Jason Sheltzer, PhD, of Cold Spring Harbor (N.Y.) Laboratory and colleagues found that the cancer cells could get along just fine without the targeted proteins, suggesting that the drugs’ alleged efficacy in the lab dish was because of other, off-target effects.
“It seemed like these genes that encode proteins that are being targeted by putative precision agents in clinical trials aren’t actually essential at all for cancer cell growth, so that was one surprise that we found,” Dr. Sheltzer said in a telephone briefing for reporters held prior to publication of the study in Science Translational Medicine.
“The second surprise was that we took the drugs that were supposed to be specific for these proteins and then we treated cancer cells with them, and we found that the drugs continued to kill the cancer cells that totally lacked the target protein expression,” he said.
But the investigators also made a serendipitous discovery that one of the drugs they tested, OTS964, was not – as originally thought – an inhibitor of the PBK protein but instead was an inhibitor of cyclin-dependent kinase (CDK) 11, making it a molecular relative of drugs such as the CDK4/6 inhibitors ribociclib (Kisqali) and palbociclib (Ibrance), both potent inhibitors of hormone receptor–positive, HER2-negative metastatic breast cancer.
Drug development insights
Their findings also indicate that less-precise candidate-drug identification techniques using RNA interference (RNAi) to knock down protein expression may have led earlier investigators down the garden path, resulting in errors that can lead to the all-too-familiar scenario of a seemingly promising compound flourishing in the early drug development process, only to wither on the vine in clinical trials.
“Everyone knows that it’s really hard to make new cancer drugs, but what we’ve been finding out is that, once a new drug is even made, it can be just as difficult to really understand how that drug is working to kill cancer cells,” said coauthor Chris Giuliano, currently a doctoral candidate at the Massachusetts Institute of Technology in Cambridge, who also spoke at the briefing.
“Our study showed us that a potential problem with the cancer drug development pipeline is that the way in which some of these new cancer drugs work is incompletely understood. Ten years ago many of these studies were developed with a tool known as RNAi, which while being the best available tool at the time ultimately led many researchers to arrive at the wrong conclusions about how some of these drug targets actually work,” he added.
Sour on MELK
The investigators had previously found that MELK (maternal embryonic leucine zipper kinase), a protein previously identified as essential to survival in multiple cancer types, could be eliminated from cancer cells using CRISPR gene editing without significant harm to the cells, and that a drug targeted against MELK in phase 2 clinical trials (OTS167) continued to kill the knockout cells in a lab dish with no loss of potency. This finding alerted the researchers to the possibility that drugs in development could be targeting the wrong protein, accounting for at least some of the high failure rate in cancer drug development, and potentially explaining some of the toxicities seen with experimental agents.
“Moreover, clinical trials that use a biomarker to select patients for trial inclusion are about twice as likely to succeed as those without one. Misidentifying a drug’s mechanism of action could hamper efforts to uncover a biomarker capable of predicting therapeutic responses, further decreasing the success rate of clinical trials,” they wrote.
Other false targets
In the current study, the investigators tested whether other drugs were designed to point toward nonessential or “superfluous” targets, and whether the mechanisms of action of the drugs had been mischaracterized.
They focused on five proteins that were thought to be so important to cancer cells that their loss would inhibit or block cell proliferation (HDAC6, MAPK14/p38, PAK4, PBK, and PIM1) and one (CASP3/caspase-3) that was thought to induce apoptosis when activated by a small molecule.
First, the investigators determined that the putative targets – the five proteins listed before – may not be required for actual cancer cell growth or survival, and then found evidence to suggest that misidentification of the proteins may have been caused by the uncertainties of RNAi.
They then used CRISPR to assess the mechanism of action of each of the 10 drugs, and whether the effects they induced were on or off target. They found that PAC-1, a putative caspase-3 activator currently in three clinical trials, actually works in a caspase-3–independent manner, and that all 10 anticancer drugs “exhibited clear evidence of target-independent cell killing in every [knockout] cell line that we examined.”
Finally, as noted before, they determined that the actual mechanism of action of OTS964 was not PBK inhibition, but inhibition of CDK11, a protein that appears to be vital for mitosis in human cancers.
“We think that CDK11 is an exciting target for future therapeutic development, and we found it specifically by looking for the true targets of these mischaracterized agents,” Dr. Sheltzer said.
The investigators acknowledged that their study was limited by the use of well-established cancer cell lines that may not fully reflect how cancer acts in the human body, and they could not rule out that the superfluous proteins they identified might be important for the survival of rare cancers.
“Additionally, we’re not saying that these targets offer no therapeutic potential,” said lead author Ann Lin, who is currently a Fulbright Fellow at the University of Oslo.
“It might be that there are other, unrelated proteins in the cell taking over its role and targeting of both proteins in combination is needed to kill the cancer cells. Furthermore, removal of these proteins may reveal a weakness in the cancer that can be targeted by a second drug, so our experiments showed that uniquely targeting these proteins alone showed little efficacy,” she said.
Research in the Sheltzer Lab is supported by an National Institutes of Health Early Independence Award, a Breast Cancer Alliance Young Investigator Award, a Damon Runyon-Rachleff Innovation Award, a Gates Foundation Innovative Technology Solutions grant, and a CSHL-Northwell Translational Cancer Research grant. The authors reported that they have no competing interests.
SOURCE: Lin A et al. Sci Transl Med. 2019 Sep 11. doi: 10.1126/scitranslmed.aaw8412 .
Clinical trials of novel cancer drugs miss their marks far more often than they hit them, in part because the drugs themselves may be aimed at the wrong targets or the targets themselves may not be that important in the first place, investigators have found.
Using CRISPR (clustered regularly interspaced palindromic repeats) gene editing to study the effects of 10 drugs that are in development targeting six proteins ostensibly crucial to the health or survival of cancer cells, Jason Sheltzer, PhD, of Cold Spring Harbor (N.Y.) Laboratory and colleagues found that the cancer cells could get along just fine without the targeted proteins, suggesting that the drugs’ alleged efficacy in the lab dish was because of other, off-target effects.
“It seemed like these genes that encode proteins that are being targeted by putative precision agents in clinical trials aren’t actually essential at all for cancer cell growth, so that was one surprise that we found,” Dr. Sheltzer said in a telephone briefing for reporters held prior to publication of the study in Science Translational Medicine.
“The second surprise was that we took the drugs that were supposed to be specific for these proteins and then we treated cancer cells with them, and we found that the drugs continued to kill the cancer cells that totally lacked the target protein expression,” he said.
But the investigators also made a serendipitous discovery that one of the drugs they tested, OTS964, was not – as originally thought – an inhibitor of the PBK protein but instead was an inhibitor of cyclin-dependent kinase (CDK) 11, making it a molecular relative of drugs such as the CDK4/6 inhibitors ribociclib (Kisqali) and palbociclib (Ibrance), both potent inhibitors of hormone receptor–positive, HER2-negative metastatic breast cancer.
Drug development insights
Their findings also indicate that less-precise candidate-drug identification techniques using RNA interference (RNAi) to knock down protein expression may have led earlier investigators down the garden path, resulting in errors that can lead to the all-too-familiar scenario of a seemingly promising compound flourishing in the early drug development process, only to wither on the vine in clinical trials.
“Everyone knows that it’s really hard to make new cancer drugs, but what we’ve been finding out is that, once a new drug is even made, it can be just as difficult to really understand how that drug is working to kill cancer cells,” said coauthor Chris Giuliano, currently a doctoral candidate at the Massachusetts Institute of Technology in Cambridge, who also spoke at the briefing.
“Our study showed us that a potential problem with the cancer drug development pipeline is that the way in which some of these new cancer drugs work is incompletely understood. Ten years ago many of these studies were developed with a tool known as RNAi, which while being the best available tool at the time ultimately led many researchers to arrive at the wrong conclusions about how some of these drug targets actually work,” he added.
Sour on MELK
The investigators had previously found that MELK (maternal embryonic leucine zipper kinase), a protein previously identified as essential to survival in multiple cancer types, could be eliminated from cancer cells using CRISPR gene editing without significant harm to the cells, and that a drug targeted against MELK in phase 2 clinical trials (OTS167) continued to kill the knockout cells in a lab dish with no loss of potency. This finding alerted the researchers to the possibility that drugs in development could be targeting the wrong protein, accounting for at least some of the high failure rate in cancer drug development, and potentially explaining some of the toxicities seen with experimental agents.
“Moreover, clinical trials that use a biomarker to select patients for trial inclusion are about twice as likely to succeed as those without one. Misidentifying a drug’s mechanism of action could hamper efforts to uncover a biomarker capable of predicting therapeutic responses, further decreasing the success rate of clinical trials,” they wrote.
Other false targets
In the current study, the investigators tested whether other drugs were designed to point toward nonessential or “superfluous” targets, and whether the mechanisms of action of the drugs had been mischaracterized.
They focused on five proteins that were thought to be so important to cancer cells that their loss would inhibit or block cell proliferation (HDAC6, MAPK14/p38, PAK4, PBK, and PIM1) and one (CASP3/caspase-3) that was thought to induce apoptosis when activated by a small molecule.
First, the investigators determined that the putative targets – the five proteins listed before – may not be required for actual cancer cell growth or survival, and then found evidence to suggest that misidentification of the proteins may have been caused by the uncertainties of RNAi.
They then used CRISPR to assess the mechanism of action of each of the 10 drugs, and whether the effects they induced were on or off target. They found that PAC-1, a putative caspase-3 activator currently in three clinical trials, actually works in a caspase-3–independent manner, and that all 10 anticancer drugs “exhibited clear evidence of target-independent cell killing in every [knockout] cell line that we examined.”
Finally, as noted before, they determined that the actual mechanism of action of OTS964 was not PBK inhibition, but inhibition of CDK11, a protein that appears to be vital for mitosis in human cancers.
“We think that CDK11 is an exciting target for future therapeutic development, and we found it specifically by looking for the true targets of these mischaracterized agents,” Dr. Sheltzer said.
The investigators acknowledged that their study was limited by the use of well-established cancer cell lines that may not fully reflect how cancer acts in the human body, and they could not rule out that the superfluous proteins they identified might be important for the survival of rare cancers.
“Additionally, we’re not saying that these targets offer no therapeutic potential,” said lead author Ann Lin, who is currently a Fulbright Fellow at the University of Oslo.
“It might be that there are other, unrelated proteins in the cell taking over its role and targeting of both proteins in combination is needed to kill the cancer cells. Furthermore, removal of these proteins may reveal a weakness in the cancer that can be targeted by a second drug, so our experiments showed that uniquely targeting these proteins alone showed little efficacy,” she said.
Research in the Sheltzer Lab is supported by an National Institutes of Health Early Independence Award, a Breast Cancer Alliance Young Investigator Award, a Damon Runyon-Rachleff Innovation Award, a Gates Foundation Innovative Technology Solutions grant, and a CSHL-Northwell Translational Cancer Research grant. The authors reported that they have no competing interests.
SOURCE: Lin A et al. Sci Transl Med. 2019 Sep 11. doi: 10.1126/scitranslmed.aaw8412 .
Clinical trials of novel cancer drugs miss their marks far more often than they hit them, in part because the drugs themselves may be aimed at the wrong targets or the targets themselves may not be that important in the first place, investigators have found.
Using CRISPR (clustered regularly interspaced palindromic repeats) gene editing to study the effects of 10 drugs that are in development targeting six proteins ostensibly crucial to the health or survival of cancer cells, Jason Sheltzer, PhD, of Cold Spring Harbor (N.Y.) Laboratory and colleagues found that the cancer cells could get along just fine without the targeted proteins, suggesting that the drugs’ alleged efficacy in the lab dish was because of other, off-target effects.
“It seemed like these genes that encode proteins that are being targeted by putative precision agents in clinical trials aren’t actually essential at all for cancer cell growth, so that was one surprise that we found,” Dr. Sheltzer said in a telephone briefing for reporters held prior to publication of the study in Science Translational Medicine.
“The second surprise was that we took the drugs that were supposed to be specific for these proteins and then we treated cancer cells with them, and we found that the drugs continued to kill the cancer cells that totally lacked the target protein expression,” he said.
But the investigators also made a serendipitous discovery that one of the drugs they tested, OTS964, was not – as originally thought – an inhibitor of the PBK protein but instead was an inhibitor of cyclin-dependent kinase (CDK) 11, making it a molecular relative of drugs such as the CDK4/6 inhibitors ribociclib (Kisqali) and palbociclib (Ibrance), both potent inhibitors of hormone receptor–positive, HER2-negative metastatic breast cancer.
Drug development insights
Their findings also indicate that less-precise candidate-drug identification techniques using RNA interference (RNAi) to knock down protein expression may have led earlier investigators down the garden path, resulting in errors that can lead to the all-too-familiar scenario of a seemingly promising compound flourishing in the early drug development process, only to wither on the vine in clinical trials.
“Everyone knows that it’s really hard to make new cancer drugs, but what we’ve been finding out is that, once a new drug is even made, it can be just as difficult to really understand how that drug is working to kill cancer cells,” said coauthor Chris Giuliano, currently a doctoral candidate at the Massachusetts Institute of Technology in Cambridge, who also spoke at the briefing.
“Our study showed us that a potential problem with the cancer drug development pipeline is that the way in which some of these new cancer drugs work is incompletely understood. Ten years ago many of these studies were developed with a tool known as RNAi, which while being the best available tool at the time ultimately led many researchers to arrive at the wrong conclusions about how some of these drug targets actually work,” he added.
Sour on MELK
The investigators had previously found that MELK (maternal embryonic leucine zipper kinase), a protein previously identified as essential to survival in multiple cancer types, could be eliminated from cancer cells using CRISPR gene editing without significant harm to the cells, and that a drug targeted against MELK in phase 2 clinical trials (OTS167) continued to kill the knockout cells in a lab dish with no loss of potency. This finding alerted the researchers to the possibility that drugs in development could be targeting the wrong protein, accounting for at least some of the high failure rate in cancer drug development, and potentially explaining some of the toxicities seen with experimental agents.
“Moreover, clinical trials that use a biomarker to select patients for trial inclusion are about twice as likely to succeed as those without one. Misidentifying a drug’s mechanism of action could hamper efforts to uncover a biomarker capable of predicting therapeutic responses, further decreasing the success rate of clinical trials,” they wrote.
Other false targets
In the current study, the investigators tested whether other drugs were designed to point toward nonessential or “superfluous” targets, and whether the mechanisms of action of the drugs had been mischaracterized.
They focused on five proteins that were thought to be so important to cancer cells that their loss would inhibit or block cell proliferation (HDAC6, MAPK14/p38, PAK4, PBK, and PIM1) and one (CASP3/caspase-3) that was thought to induce apoptosis when activated by a small molecule.
First, the investigators determined that the putative targets – the five proteins listed before – may not be required for actual cancer cell growth or survival, and then found evidence to suggest that misidentification of the proteins may have been caused by the uncertainties of RNAi.
They then used CRISPR to assess the mechanism of action of each of the 10 drugs, and whether the effects they induced were on or off target. They found that PAC-1, a putative caspase-3 activator currently in three clinical trials, actually works in a caspase-3–independent manner, and that all 10 anticancer drugs “exhibited clear evidence of target-independent cell killing in every [knockout] cell line that we examined.”
Finally, as noted before, they determined that the actual mechanism of action of OTS964 was not PBK inhibition, but inhibition of CDK11, a protein that appears to be vital for mitosis in human cancers.
“We think that CDK11 is an exciting target for future therapeutic development, and we found it specifically by looking for the true targets of these mischaracterized agents,” Dr. Sheltzer said.
The investigators acknowledged that their study was limited by the use of well-established cancer cell lines that may not fully reflect how cancer acts in the human body, and they could not rule out that the superfluous proteins they identified might be important for the survival of rare cancers.
“Additionally, we’re not saying that these targets offer no therapeutic potential,” said lead author Ann Lin, who is currently a Fulbright Fellow at the University of Oslo.
“It might be that there are other, unrelated proteins in the cell taking over its role and targeting of both proteins in combination is needed to kill the cancer cells. Furthermore, removal of these proteins may reveal a weakness in the cancer that can be targeted by a second drug, so our experiments showed that uniquely targeting these proteins alone showed little efficacy,” she said.
Research in the Sheltzer Lab is supported by an National Institutes of Health Early Independence Award, a Breast Cancer Alliance Young Investigator Award, a Damon Runyon-Rachleff Innovation Award, a Gates Foundation Innovative Technology Solutions grant, and a CSHL-Northwell Translational Cancer Research grant. The authors reported that they have no competing interests.
SOURCE: Lin A et al. Sci Transl Med. 2019 Sep 11. doi: 10.1126/scitranslmed.aaw8412 .
FROM SCIENCE TRANSLATIONAL MEDICINE
Key clinical point: Cancer drug developers need more stringent methods for verifying targets before advancing drugs to clinical trials.
Major finding: Ten drugs in development against cancer have mechanisms of action independent of their targets.
Study details: In vitro studies of cancer drugs and their targets using CRISPR gene-editing techniques.
Disclosures: Research in the Sheltzer Lab is supported by a National Institutes of Health Early Independence Award, a Breast Cancer Alliance Young Investigator Award, a Damon Runyon-Rachleff Innovation Award, a Gates Foundation Innovative Technology Solutions grant, and a CSHL-Northwell Translational Cancer Research grant. The authors reported that they have no competing interests.
Source: Lin A et al. Sci Transl Med. 2019 Sep 11. doi: 10.1126/scitranslmed.aaw8412.
Tissue TMB disappoints as treatment response biomarker in NSCLC
BARCELONA – Tissue tumor mutational burden (TMB) was not significantly associated with treatment efficacy in patients with metastatic nonsquamous non–small cell lung cancer (NSCLC) in the phase 3 KEYNOTE-189 study and the phase 2 KEYNOTE-021 study.
In 293 patients with evaluable TMB data in KEYNOTE-189, including 207 who were treated with pembrolizumab plus chemotherapy and 86 who received placebo plus chemotherapy, TMB as a continuous variable showed no significant association with either overall survival (OS), progression-free survival (PFS), or objective response rate (ORR), Marina C. Garassino, MD, of Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, reported at the World Conference on Lung Cancer.
Pembrolizumab plus chemotherapy improved OS both in patients with TMB of 175 mutations/exome and greater and in those with TMB of fewer than 175 mutations/exome (hazard ratio for OS, 0.64 for both), and similar results were seen for PFS and ORR, Dr. Garassino said at the conference, which is sponsored by the International Association for the Study of Lung Cancer. Similar results were also seen for those with tissue TMB of 150 mutations/exome or greater and those with fewer than 150 mutations/exome, she noted.
The double-blind KEYNOTE-189 study compared first-line pembrolizumab plus chemotherapy with placebo plus chemotherapy in 616 patients who were randomized 2:1 to the treatment arms, respectively, and showed that adding pembrolizumab to pemetrexed and platinum significantly improved OS (HR, 0.49), PFS (HR, 0.52), and ORR (47.6% vs. 18.9%). Benefit was observed in all analyzed subgroups, including patients with programmed death-ligand 1 (PD-L1) Tissue Polypeptide-specific (TPS) antigen of less than 1%, 1-49%, and 50% or greater, she noted.
In the current analysis, performed to assess the effect of tissue TMB on response rates, a similar benefit was seen in both TMB-high and -low subgroups.
“Our data suggest that tissue TMB may not help select patients who would have better outcomes with pembrolizumab plus pemetrexed and a platinum given as first-line therapy for metastatic nonsquamous NSCLC,” she concluded.
Similarly, an exploratory analysis of data from the open-label, phase 2 KEYNOTE-021 trial – the first trial to show the efficacy and safety of the anti–PD-1 immune checkpoint inhibitor pembrolizumab given with chemotherapy – showed no association between tissue TMB and OS, PFS, or ORR in 70 patients with metastatic nonsquamous NSCLC who were treated with either pembrolizumab plus carboplatin and pemetrexed or with carboplatin and pemetrexed alone, Corey Langer, MD, reported at the conference.
“As you’re well aware,TMB has been widely evaluated as a biomarker for immunotherapy in advanced [NSCLC] and may identify patients who are more likely to respond to immune checkpoint inhibitors,” said Dr. Langer, professor of medicine and director of thoracic surgery at the Hospital of the University of Pennsylvania, Philadelphia. “But we have very limited data on whether TMB is of any value as a biomarker for chemo, either alone or given with an immune checkpoint inhibitor.”
In this analysis, pembrolizumab plus chemotherapy was associated with a high response rate, regardless of tissue TMB status; in those with tissue TMB of 175 mutations/exome or greater and fewer than 175 mutations/exome, the response rates were 71% and 61%, respectively.
“So, tissue TMB assessed by whole-exome sequencing was not significantly associated with efficacy for pembro and combination pem-carbo, or for chemotherapy alone, for first-line treatment of patients with metastatic nonsquamous [NSCLC], nor was there any significant association with PD-L1 expression,” he said. “Obviously an analysis of much larger datasets is needed to assess whether the benefit of pembro plus chemo relative to chemo alone differs in patients with TMB-high or TMB-low tumors,” he said.
For now, tissue TMB should not be used – “at least not yet” – in therapeutic decision making, he said, adding that it is important to distinguish between blood TMB and tissue TMB because the latter “may be more reflective of the entire tumor.”
Dr. Langer noted that he still thinks TMB has a potential role.
“We just haven’t figured it out yet,” he said.
Both KEYNOTE-189 and KEYNOTE-021 were supported by Merck. Dr. Garassino and Dr. Langer reported relationships with several pharmaceutical companies.
SOURCES: Garassino MC et al. WCLC 2019, Abstract OA04.06; Langer C et al. WCLC 2019, Abstract OA04.05.
.
BARCELONA – Tissue tumor mutational burden (TMB) was not significantly associated with treatment efficacy in patients with metastatic nonsquamous non–small cell lung cancer (NSCLC) in the phase 3 KEYNOTE-189 study and the phase 2 KEYNOTE-021 study.
In 293 patients with evaluable TMB data in KEYNOTE-189, including 207 who were treated with pembrolizumab plus chemotherapy and 86 who received placebo plus chemotherapy, TMB as a continuous variable showed no significant association with either overall survival (OS), progression-free survival (PFS), or objective response rate (ORR), Marina C. Garassino, MD, of Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, reported at the World Conference on Lung Cancer.
Pembrolizumab plus chemotherapy improved OS both in patients with TMB of 175 mutations/exome and greater and in those with TMB of fewer than 175 mutations/exome (hazard ratio for OS, 0.64 for both), and similar results were seen for PFS and ORR, Dr. Garassino said at the conference, which is sponsored by the International Association for the Study of Lung Cancer. Similar results were also seen for those with tissue TMB of 150 mutations/exome or greater and those with fewer than 150 mutations/exome, she noted.
The double-blind KEYNOTE-189 study compared first-line pembrolizumab plus chemotherapy with placebo plus chemotherapy in 616 patients who were randomized 2:1 to the treatment arms, respectively, and showed that adding pembrolizumab to pemetrexed and platinum significantly improved OS (HR, 0.49), PFS (HR, 0.52), and ORR (47.6% vs. 18.9%). Benefit was observed in all analyzed subgroups, including patients with programmed death-ligand 1 (PD-L1) Tissue Polypeptide-specific (TPS) antigen of less than 1%, 1-49%, and 50% or greater, she noted.
In the current analysis, performed to assess the effect of tissue TMB on response rates, a similar benefit was seen in both TMB-high and -low subgroups.
“Our data suggest that tissue TMB may not help select patients who would have better outcomes with pembrolizumab plus pemetrexed and a platinum given as first-line therapy for metastatic nonsquamous NSCLC,” she concluded.
Similarly, an exploratory analysis of data from the open-label, phase 2 KEYNOTE-021 trial – the first trial to show the efficacy and safety of the anti–PD-1 immune checkpoint inhibitor pembrolizumab given with chemotherapy – showed no association between tissue TMB and OS, PFS, or ORR in 70 patients with metastatic nonsquamous NSCLC who were treated with either pembrolizumab plus carboplatin and pemetrexed or with carboplatin and pemetrexed alone, Corey Langer, MD, reported at the conference.
“As you’re well aware,TMB has been widely evaluated as a biomarker for immunotherapy in advanced [NSCLC] and may identify patients who are more likely to respond to immune checkpoint inhibitors,” said Dr. Langer, professor of medicine and director of thoracic surgery at the Hospital of the University of Pennsylvania, Philadelphia. “But we have very limited data on whether TMB is of any value as a biomarker for chemo, either alone or given with an immune checkpoint inhibitor.”
In this analysis, pembrolizumab plus chemotherapy was associated with a high response rate, regardless of tissue TMB status; in those with tissue TMB of 175 mutations/exome or greater and fewer than 175 mutations/exome, the response rates were 71% and 61%, respectively.
“So, tissue TMB assessed by whole-exome sequencing was not significantly associated with efficacy for pembro and combination pem-carbo, or for chemotherapy alone, for first-line treatment of patients with metastatic nonsquamous [NSCLC], nor was there any significant association with PD-L1 expression,” he said. “Obviously an analysis of much larger datasets is needed to assess whether the benefit of pembro plus chemo relative to chemo alone differs in patients with TMB-high or TMB-low tumors,” he said.
For now, tissue TMB should not be used – “at least not yet” – in therapeutic decision making, he said, adding that it is important to distinguish between blood TMB and tissue TMB because the latter “may be more reflective of the entire tumor.”
Dr. Langer noted that he still thinks TMB has a potential role.
“We just haven’t figured it out yet,” he said.
Both KEYNOTE-189 and KEYNOTE-021 were supported by Merck. Dr. Garassino and Dr. Langer reported relationships with several pharmaceutical companies.
SOURCES: Garassino MC et al. WCLC 2019, Abstract OA04.06; Langer C et al. WCLC 2019, Abstract OA04.05.
.
BARCELONA – Tissue tumor mutational burden (TMB) was not significantly associated with treatment efficacy in patients with metastatic nonsquamous non–small cell lung cancer (NSCLC) in the phase 3 KEYNOTE-189 study and the phase 2 KEYNOTE-021 study.
In 293 patients with evaluable TMB data in KEYNOTE-189, including 207 who were treated with pembrolizumab plus chemotherapy and 86 who received placebo plus chemotherapy, TMB as a continuous variable showed no significant association with either overall survival (OS), progression-free survival (PFS), or objective response rate (ORR), Marina C. Garassino, MD, of Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, reported at the World Conference on Lung Cancer.
Pembrolizumab plus chemotherapy improved OS both in patients with TMB of 175 mutations/exome and greater and in those with TMB of fewer than 175 mutations/exome (hazard ratio for OS, 0.64 for both), and similar results were seen for PFS and ORR, Dr. Garassino said at the conference, which is sponsored by the International Association for the Study of Lung Cancer. Similar results were also seen for those with tissue TMB of 150 mutations/exome or greater and those with fewer than 150 mutations/exome, she noted.
The double-blind KEYNOTE-189 study compared first-line pembrolizumab plus chemotherapy with placebo plus chemotherapy in 616 patients who were randomized 2:1 to the treatment arms, respectively, and showed that adding pembrolizumab to pemetrexed and platinum significantly improved OS (HR, 0.49), PFS (HR, 0.52), and ORR (47.6% vs. 18.9%). Benefit was observed in all analyzed subgroups, including patients with programmed death-ligand 1 (PD-L1) Tissue Polypeptide-specific (TPS) antigen of less than 1%, 1-49%, and 50% or greater, she noted.
In the current analysis, performed to assess the effect of tissue TMB on response rates, a similar benefit was seen in both TMB-high and -low subgroups.
“Our data suggest that tissue TMB may not help select patients who would have better outcomes with pembrolizumab plus pemetrexed and a platinum given as first-line therapy for metastatic nonsquamous NSCLC,” she concluded.
Similarly, an exploratory analysis of data from the open-label, phase 2 KEYNOTE-021 trial – the first trial to show the efficacy and safety of the anti–PD-1 immune checkpoint inhibitor pembrolizumab given with chemotherapy – showed no association between tissue TMB and OS, PFS, or ORR in 70 patients with metastatic nonsquamous NSCLC who were treated with either pembrolizumab plus carboplatin and pemetrexed or with carboplatin and pemetrexed alone, Corey Langer, MD, reported at the conference.
“As you’re well aware,TMB has been widely evaluated as a biomarker for immunotherapy in advanced [NSCLC] and may identify patients who are more likely to respond to immune checkpoint inhibitors,” said Dr. Langer, professor of medicine and director of thoracic surgery at the Hospital of the University of Pennsylvania, Philadelphia. “But we have very limited data on whether TMB is of any value as a biomarker for chemo, either alone or given with an immune checkpoint inhibitor.”
In this analysis, pembrolizumab plus chemotherapy was associated with a high response rate, regardless of tissue TMB status; in those with tissue TMB of 175 mutations/exome or greater and fewer than 175 mutations/exome, the response rates were 71% and 61%, respectively.
“So, tissue TMB assessed by whole-exome sequencing was not significantly associated with efficacy for pembro and combination pem-carbo, or for chemotherapy alone, for first-line treatment of patients with metastatic nonsquamous [NSCLC], nor was there any significant association with PD-L1 expression,” he said. “Obviously an analysis of much larger datasets is needed to assess whether the benefit of pembro plus chemo relative to chemo alone differs in patients with TMB-high or TMB-low tumors,” he said.
For now, tissue TMB should not be used – “at least not yet” – in therapeutic decision making, he said, adding that it is important to distinguish between blood TMB and tissue TMB because the latter “may be more reflective of the entire tumor.”
Dr. Langer noted that he still thinks TMB has a potential role.
“We just haven’t figured it out yet,” he said.
Both KEYNOTE-189 and KEYNOTE-021 were supported by Merck. Dr. Garassino and Dr. Langer reported relationships with several pharmaceutical companies.
SOURCES: Garassino MC et al. WCLC 2019, Abstract OA04.06; Langer C et al. WCLC 2019, Abstract OA04.05.
.
REPORTING FROM WCLC 2019
Use of genetic testing for congenital heart defect management
The average student in America learns that genes form the building blocks of what makes us human by the time they receive their high school diploma. Indeed, the completion of the Human Genome Project in 2003 paved the way for our genetic makeup, much like our medical history, to become a routine part of our health care. For example, our faculty at the University of Maryland School of Medicine discovered an important gene – CYP2C19 – which is involved in the metabolism of the antiplatelet medicine clopidogrel (Plavix). Although most people have this gene, some don’t. Therefore, when we manage a patient with coronary disease, we use a genetic screen to determine whether that patient has CYP2C19 and then modify therapy based on these results.
Our genes also have become commodities – from companies willing to analyze our genes to determine our racial and ethnic ancestry or propensity for certain diseases to those that can sequence the family dog’s genes.
Advances in genomics similarly have impacted ob.gyn. practice. Because of rapidly evolving gene analysis tools, we can now, for example, noninvasively test a developing fetus’s risk for chromosomal abnormalities and determine a baby’s sex by merely examining fetal DNA in a pregnant woman’s bloodstream. Although not diagnostic, these gene-based prenatal screening tests have reduced the need for unnecessary, costly, and highly invasive procedures for many of our patients.
Importantly, our recognition that certain genes can confer a higher risk of disease has meant that performing a prenatal genetic evaluation can greatly inform the mother and her care team about potential problems her baby may have that may require additional management. For babies who have congenital heart defects, a genetic evaluation performed in addition to sonographic examination can provide ob.gyns. with crucial details to enhance pregnancy management and postnatal care decisions.
The importance of genetic testing and analysis in the detection, treatment, and prevention of congenital heart defects is the topic of part two of this two-part Master Class series authored by Shifa Turan, MD, associate professor of obstetrics, gynecology, and reproductive sciences at the University of Maryland School of Medicine and director of the Fetal Heart Program at the University of Maryland Medical Center. By using a combination of three- and four-dimensional ultrasound with gene assays, Dr. Turan and her colleagues can greatly enhance and personalize the care they deliver to their patients.
Dr. Reece, who specializes in maternal-fetal medicine, is executive vice president for medical affairs at the University of Maryland School of Medicine, as well as the John Z. and Akiko K. Bowers Distinguished Professor and dean of the school of medicine. He is the medical editor of this column. He said he had no relevant financial disclosures. Contact him at obnews@mdedge.com.
The average student in America learns that genes form the building blocks of what makes us human by the time they receive their high school diploma. Indeed, the completion of the Human Genome Project in 2003 paved the way for our genetic makeup, much like our medical history, to become a routine part of our health care. For example, our faculty at the University of Maryland School of Medicine discovered an important gene – CYP2C19 – which is involved in the metabolism of the antiplatelet medicine clopidogrel (Plavix). Although most people have this gene, some don’t. Therefore, when we manage a patient with coronary disease, we use a genetic screen to determine whether that patient has CYP2C19 and then modify therapy based on these results.
Our genes also have become commodities – from companies willing to analyze our genes to determine our racial and ethnic ancestry or propensity for certain diseases to those that can sequence the family dog’s genes.
Advances in genomics similarly have impacted ob.gyn. practice. Because of rapidly evolving gene analysis tools, we can now, for example, noninvasively test a developing fetus’s risk for chromosomal abnormalities and determine a baby’s sex by merely examining fetal DNA in a pregnant woman’s bloodstream. Although not diagnostic, these gene-based prenatal screening tests have reduced the need for unnecessary, costly, and highly invasive procedures for many of our patients.
Importantly, our recognition that certain genes can confer a higher risk of disease has meant that performing a prenatal genetic evaluation can greatly inform the mother and her care team about potential problems her baby may have that may require additional management. For babies who have congenital heart defects, a genetic evaluation performed in addition to sonographic examination can provide ob.gyns. with crucial details to enhance pregnancy management and postnatal care decisions.
The importance of genetic testing and analysis in the detection, treatment, and prevention of congenital heart defects is the topic of part two of this two-part Master Class series authored by Shifa Turan, MD, associate professor of obstetrics, gynecology, and reproductive sciences at the University of Maryland School of Medicine and director of the Fetal Heart Program at the University of Maryland Medical Center. By using a combination of three- and four-dimensional ultrasound with gene assays, Dr. Turan and her colleagues can greatly enhance and personalize the care they deliver to their patients.
Dr. Reece, who specializes in maternal-fetal medicine, is executive vice president for medical affairs at the University of Maryland School of Medicine, as well as the John Z. and Akiko K. Bowers Distinguished Professor and dean of the school of medicine. He is the medical editor of this column. He said he had no relevant financial disclosures. Contact him at obnews@mdedge.com.
The average student in America learns that genes form the building blocks of what makes us human by the time they receive their high school diploma. Indeed, the completion of the Human Genome Project in 2003 paved the way for our genetic makeup, much like our medical history, to become a routine part of our health care. For example, our faculty at the University of Maryland School of Medicine discovered an important gene – CYP2C19 – which is involved in the metabolism of the antiplatelet medicine clopidogrel (Plavix). Although most people have this gene, some don’t. Therefore, when we manage a patient with coronary disease, we use a genetic screen to determine whether that patient has CYP2C19 and then modify therapy based on these results.
Our genes also have become commodities – from companies willing to analyze our genes to determine our racial and ethnic ancestry or propensity for certain diseases to those that can sequence the family dog’s genes.
Advances in genomics similarly have impacted ob.gyn. practice. Because of rapidly evolving gene analysis tools, we can now, for example, noninvasively test a developing fetus’s risk for chromosomal abnormalities and determine a baby’s sex by merely examining fetal DNA in a pregnant woman’s bloodstream. Although not diagnostic, these gene-based prenatal screening tests have reduced the need for unnecessary, costly, and highly invasive procedures for many of our patients.
Importantly, our recognition that certain genes can confer a higher risk of disease has meant that performing a prenatal genetic evaluation can greatly inform the mother and her care team about potential problems her baby may have that may require additional management. For babies who have congenital heart defects, a genetic evaluation performed in addition to sonographic examination can provide ob.gyns. with crucial details to enhance pregnancy management and postnatal care decisions.
The importance of genetic testing and analysis in the detection, treatment, and prevention of congenital heart defects is the topic of part two of this two-part Master Class series authored by Shifa Turan, MD, associate professor of obstetrics, gynecology, and reproductive sciences at the University of Maryland School of Medicine and director of the Fetal Heart Program at the University of Maryland Medical Center. By using a combination of three- and four-dimensional ultrasound with gene assays, Dr. Turan and her colleagues can greatly enhance and personalize the care they deliver to their patients.
Dr. Reece, who specializes in maternal-fetal medicine, is executive vice president for medical affairs at the University of Maryland School of Medicine, as well as the John Z. and Akiko K. Bowers Distinguished Professor and dean of the school of medicine. He is the medical editor of this column. He said he had no relevant financial disclosures. Contact him at obnews@mdedge.com.
Genetic assessment for CHD: Case-specific, stepwise
Congenital heart defects (CHDs) are etiologically heterogeneous, but in recent years it has become clear that genetics plays a larger role in the development of CHDs than was previously thought. Research has been shifting from a focus on risk – estimating the magnitude of increased risk, for instance, based on maternal or familial risk factors – to a focus on the etiology of cardiac defects.
In practice, advances in genetic testing technologies have made the underlying causes of CHDs increasingly detectable. Chromosomal microarray analysis (CMA) – technology that detects significantly more and smaller changes in the amount of chromosomal material than traditional karyotype – has been proven to increase the diagnostic yield in cases of isolated CHDs and CHDs with extracardiac anomalies. Targeted next-generation sequencing also is now available as an additional approach in selective cases, and a clinically viable option for whole-exome sequencing is fast approaching.
For researchers, genetic evaluation carries the potential to unravel remaining mysteries about underlying causes of CHDs – to provide pathological insights and identify potential therapeutic targets. Currently, about 6 % of the total pie of presumed genetic determinants of CHDs is attributed to chromosomal anomalies, 10% to copy number variants, and 12% to single-gene defects. The remaining 72% of etiology, approximately, is undetermined.
As Helen Taussig, MD, (known as the founder of pediatric cardiology) once said, common cardiac malformations occurring in otherwise “normal” individuals “must be genetic in origin.”1 Greater use of genetic testing – and in particular, of whole-exome sequencing – will drive down this “undetermined” piece of the genetics pie.
For clinicians and patients, prenatal genetic evaluation can inform clinical management, guiding decisions on the mode, timing, and location of delivery. Genetic assessments help guide the neonatal health care team in taking optimal care of the infant, and the surgeon in preparing for neonatal surgeries and postsurgical complications.
In a recent analysis of the Society of Thoracic Surgeons Congenital Heart Surgery Database, prenatal diagnosis was associated with a lower overall prevalence of major preoperative risk factors for cardiac surgery.2 Surgical outcomes themselves also have been shown to be better after the prenatal diagnosis of complex CHDs, mainly because of improvements in perioperative care.3
When genetic etiology is elucidated, the cardiologist also is better able to counsel patients about anticipated challenges – such as the propensity, with certain genetic variants of CHD, to develop neurodevelopmental delays or other cardiac complications – and to target patient follow-up. Patients also can make informed decisions about termination or continuation of a current pregnancy and about family planning in the future.
Fortunately, advances in genetics technology have paralleled technological advancements in ultrasound. As I discussed in part one of this two-part Master Class series, it is now possible to detect many major CHDs well before 16 weeks’ gestation. Checking the structure of the fetal heart at the first-trimester screening and sonography (11-14 weeks of gestation) offers the opportunity for early genetic assessment, counseling, and planning when anomalies are detected.
A personalized approach
There has been growing interest in recent years in CMA for the prenatal genetic workup of CHDs. Microarray targets chromosomal regions at a much higher resolution than traditional karyotype. Traditional karyotype assesses both changes in chromosome number as well as more subtle structural changes such as chromosomal deletions and duplications. CMA finds what traditional karyotype identifies, but in addition, it identifies much smaller, clinically relevant chromosomal deletions and duplications that are not detected by karyotype performed with or without fluorescence in-situ hybridization (FISH). FISH uses DNA probes that carry fluorescent tags to detect chromosomal DNA.
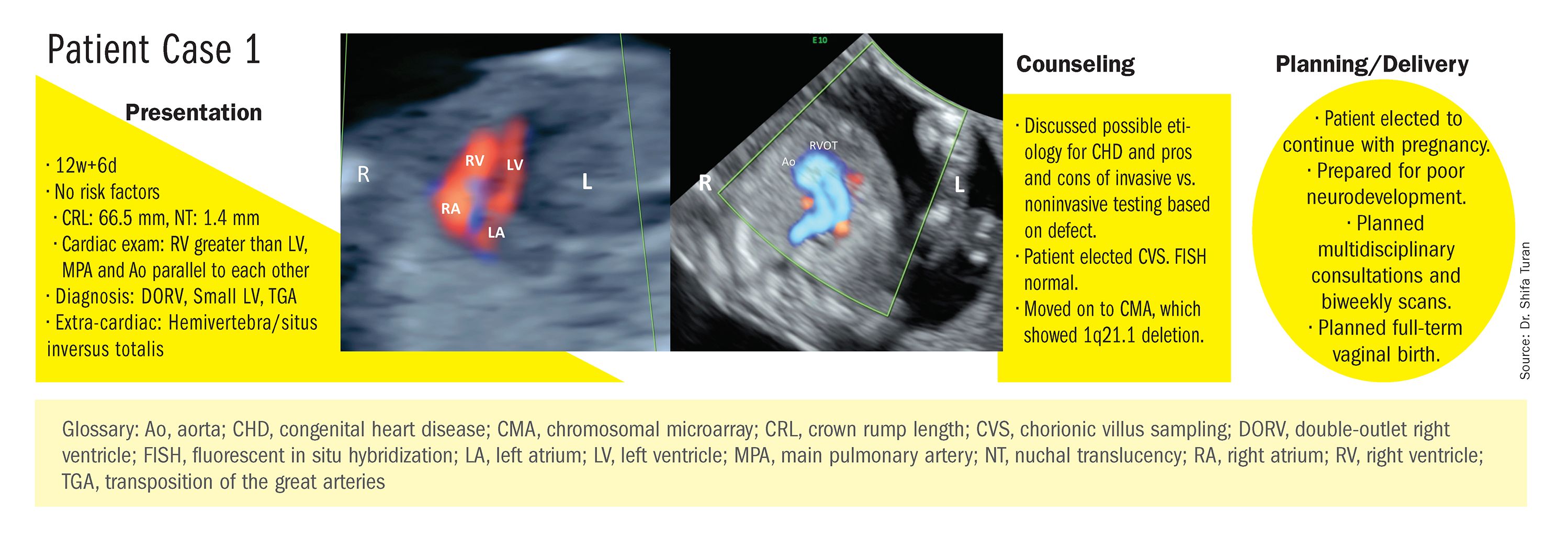
At our center, we studied the prenatal genetic test results of 145 fetuses diagnosed with CHDs. Each case involved FISH for aneuploidy/karyotype, followed by CMA in cases of a negative karyotype result. CMA increased the diagnostic yield in cases of CHD by 19.8% overall – 17.4% in cases of isolated CHD and 24.5% in cases of CHD plus extracardiac anomalies.4
Indeed, although a microarray costs more and takes an additional 2 weeks to run, CMA should be strongly considered as first-line testing for the prenatal genetic evaluation of fetuses with major structural cardiac abnormalities detected by ultrasound. However, there still are cases in which a karyotype might be sufficient. For instance, if I see that a fetus has an atrial-ventricular septal defect on a prenatal ultrasound, and there are markers for trisomy 21, 13, or 18, or Turner’s syndrome (45 XO), I usually recommend a karyotype or FISH rather than an initial CMA. If the karyotype is abnormal – which is likely in such a scenario – there isn’t a need for more extensive testing.
Similarly, when there is high suspicion for DiGeorge syndrome (the 22q11.2 deletion, which often includes cleft palate and aortic arch abnormalities), usually it is most appropriate to perform a FISH test.
CMA is the preferred first modality, however, when prenatal imaging suggests severe CHD – for instance, when there are signs of hypoplastic left heart syndrome or tetralogy of Fallot (a conotruncal defect) – or complex CHD with extracardiac anomalies. In these cases, there is a high likelihood of detecting a small deletion or duplication that would be missed with karyotype.
In the past decade, karyotype and CMA have become the major methods used in our practice. However, targeted next‐generation sequencing and whole‐exome sequencing may become more widely used because these technologies enable rapid analysis of a large number of gene sequences and facilitate discovery of novel causative genes in many genetic diseases that cause CHDs.
Currently, targeted next-generation sequencing has mainly been used in the postnatal setting, and there are limited data available on its prenatal use. Compared with whole-exome sequencing, which sequences all of the protein-coding regions of the genome, targeted next-generation sequencing panels select regions of genes that are known to be associated with diseases of interest.
For CHDs, some perinatal centers have begun using a customized gene panel that targets 77 CHD-associated genes. This particular panel has been shown to be useful in addition to current methods and is an effective tool for prenatal genetic diagnosis.5
Whole-exome sequencing is currently expensive and time consuming. While sometimes it is used in the postnatal context, it is not yet part of routine practice as a prenatal diagnostic tool. As technology advances this will change – early in the next decade, I believe. For now, whole-exome sequencing may be an option for some patients who want to know more when severe CHD is evident on ultrasound and there are negative results from CMA or targeted sequencing. We have diagnosed some rare genetic syndromes using whole-exome sequencing; these diagnoses helped us to better manage the pregnancies.
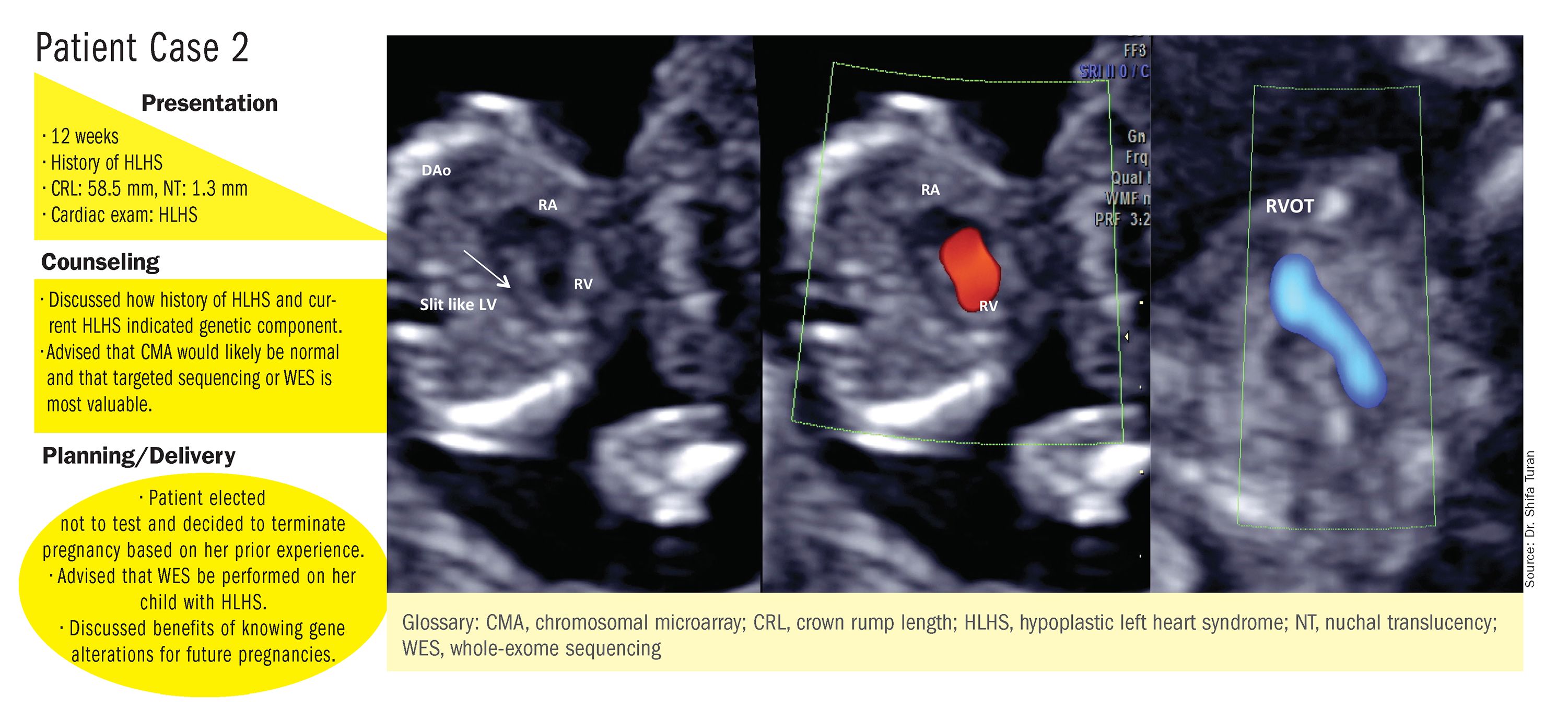
These choices are part of the case-specific, stepwise approach to genetic evaluation that we take in our fetal heart program. Our goal is to pursue information that will be accurate and valuable for the patient and clinicians, in the most cost-effective and timely manner.
Limitations of noninvasive screening
In our fetal heart program we see increasing numbers of referred patients who have chosen noninvasive cell-free fetal DNA screening (cfDNA) after a cardiac anomaly is detected on ultrasound examination, and who believe that their “low risk” results demonstrate very little or no risk of CHD. Many of these patients express a belief that noninvasive testing is highly sensitive and accurate for fetal anomalies, including CHDs, and are not easily convinced of the value of other genetic tests.
We recently conducted a retrospective chart analysis (unpublished) in which we found that 41% of cases of CHD with abnormal genetics results were not detectable by cfDNA screening.
In the case of atrial-ventricular septal defects and conotruncal abnormalities that often are more associated with common aneuploidies (trisomy 21, 18, 13, and 45 XO), a “high-risk” result from cfDNA screening may offer the family and cardiology/neonatal team some guidance, but a “low-risk” result does not eliminate the risk of a microarray abnormality and thus may provide false reassurance.
Other research has shown that noninvasive screening will miss up to 7.3% of karyotype abnormalities in pregnancies at high risk for common aneuploidies.6
While invasive testing poses a very small risk of miscarriage, it is hard without such testing to elucidate the potential genetic etiologies of CHDs and truly understand the problems. We must take time to thoughtfully counsel patients who decline invasive testing about the limitations of cfDNA screening for CHDs and other anomalies.
Dr. Turan is an associate professor of obstetrics, gynecology, and reproductive sciences, and director of the fetal heart program at the University of Maryland School of Medicine and director of the Fetal Heart Program at the University of Maryland Medical Center. Dr. Turan reported that she has no disclosures relevant to this Master Class. Email her at obnews@mdedge.com.
References
1. J Am Coll Cardiol. 1988 Oct;12(4):1079-86.
2. Pediatr Cardiol. 2019 Mar;40(3):489-96.
3. Ann Pediatr Cardiol. 2017 May-Aug;10(2):126-30.
4. Eur J Obstet Gynecol Reprod Biol 2018;221:172-76.
5. Ultrasound Obstet Gynecol. 2018 Aug;52(2):205-11.
6. PLoS One. 2016 Jan 15;11(1):e0146794.
Congenital heart defects (CHDs) are etiologically heterogeneous, but in recent years it has become clear that genetics plays a larger role in the development of CHDs than was previously thought. Research has been shifting from a focus on risk – estimating the magnitude of increased risk, for instance, based on maternal or familial risk factors – to a focus on the etiology of cardiac defects.
In practice, advances in genetic testing technologies have made the underlying causes of CHDs increasingly detectable. Chromosomal microarray analysis (CMA) – technology that detects significantly more and smaller changes in the amount of chromosomal material than traditional karyotype – has been proven to increase the diagnostic yield in cases of isolated CHDs and CHDs with extracardiac anomalies. Targeted next-generation sequencing also is now available as an additional approach in selective cases, and a clinically viable option for whole-exome sequencing is fast approaching.
For researchers, genetic evaluation carries the potential to unravel remaining mysteries about underlying causes of CHDs – to provide pathological insights and identify potential therapeutic targets. Currently, about 6 % of the total pie of presumed genetic determinants of CHDs is attributed to chromosomal anomalies, 10% to copy number variants, and 12% to single-gene defects. The remaining 72% of etiology, approximately, is undetermined.
As Helen Taussig, MD, (known as the founder of pediatric cardiology) once said, common cardiac malformations occurring in otherwise “normal” individuals “must be genetic in origin.”1 Greater use of genetic testing – and in particular, of whole-exome sequencing – will drive down this “undetermined” piece of the genetics pie.
For clinicians and patients, prenatal genetic evaluation can inform clinical management, guiding decisions on the mode, timing, and location of delivery. Genetic assessments help guide the neonatal health care team in taking optimal care of the infant, and the surgeon in preparing for neonatal surgeries and postsurgical complications.
In a recent analysis of the Society of Thoracic Surgeons Congenital Heart Surgery Database, prenatal diagnosis was associated with a lower overall prevalence of major preoperative risk factors for cardiac surgery.2 Surgical outcomes themselves also have been shown to be better after the prenatal diagnosis of complex CHDs, mainly because of improvements in perioperative care.3
When genetic etiology is elucidated, the cardiologist also is better able to counsel patients about anticipated challenges – such as the propensity, with certain genetic variants of CHD, to develop neurodevelopmental delays or other cardiac complications – and to target patient follow-up. Patients also can make informed decisions about termination or continuation of a current pregnancy and about family planning in the future.
Fortunately, advances in genetics technology have paralleled technological advancements in ultrasound. As I discussed in part one of this two-part Master Class series, it is now possible to detect many major CHDs well before 16 weeks’ gestation. Checking the structure of the fetal heart at the first-trimester screening and sonography (11-14 weeks of gestation) offers the opportunity for early genetic assessment, counseling, and planning when anomalies are detected.
A personalized approach
There has been growing interest in recent years in CMA for the prenatal genetic workup of CHDs. Microarray targets chromosomal regions at a much higher resolution than traditional karyotype. Traditional karyotype assesses both changes in chromosome number as well as more subtle structural changes such as chromosomal deletions and duplications. CMA finds what traditional karyotype identifies, but in addition, it identifies much smaller, clinically relevant chromosomal deletions and duplications that are not detected by karyotype performed with or without fluorescence in-situ hybridization (FISH). FISH uses DNA probes that carry fluorescent tags to detect chromosomal DNA.

At our center, we studied the prenatal genetic test results of 145 fetuses diagnosed with CHDs. Each case involved FISH for aneuploidy/karyotype, followed by CMA in cases of a negative karyotype result. CMA increased the diagnostic yield in cases of CHD by 19.8% overall – 17.4% in cases of isolated CHD and 24.5% in cases of CHD plus extracardiac anomalies.4
Indeed, although a microarray costs more and takes an additional 2 weeks to run, CMA should be strongly considered as first-line testing for the prenatal genetic evaluation of fetuses with major structural cardiac abnormalities detected by ultrasound. However, there still are cases in which a karyotype might be sufficient. For instance, if I see that a fetus has an atrial-ventricular septal defect on a prenatal ultrasound, and there are markers for trisomy 21, 13, or 18, or Turner’s syndrome (45 XO), I usually recommend a karyotype or FISH rather than an initial CMA. If the karyotype is abnormal – which is likely in such a scenario – there isn’t a need for more extensive testing.
Similarly, when there is high suspicion for DiGeorge syndrome (the 22q11.2 deletion, which often includes cleft palate and aortic arch abnormalities), usually it is most appropriate to perform a FISH test.
CMA is the preferred first modality, however, when prenatal imaging suggests severe CHD – for instance, when there are signs of hypoplastic left heart syndrome or tetralogy of Fallot (a conotruncal defect) – or complex CHD with extracardiac anomalies. In these cases, there is a high likelihood of detecting a small deletion or duplication that would be missed with karyotype.
In the past decade, karyotype and CMA have become the major methods used in our practice. However, targeted next‐generation sequencing and whole‐exome sequencing may become more widely used because these technologies enable rapid analysis of a large number of gene sequences and facilitate discovery of novel causative genes in many genetic diseases that cause CHDs.
Currently, targeted next-generation sequencing has mainly been used in the postnatal setting, and there are limited data available on its prenatal use. Compared with whole-exome sequencing, which sequences all of the protein-coding regions of the genome, targeted next-generation sequencing panels select regions of genes that are known to be associated with diseases of interest.
For CHDs, some perinatal centers have begun using a customized gene panel that targets 77 CHD-associated genes. This particular panel has been shown to be useful in addition to current methods and is an effective tool for prenatal genetic diagnosis.5
Whole-exome sequencing is currently expensive and time consuming. While sometimes it is used in the postnatal context, it is not yet part of routine practice as a prenatal diagnostic tool. As technology advances this will change – early in the next decade, I believe. For now, whole-exome sequencing may be an option for some patients who want to know more when severe CHD is evident on ultrasound and there are negative results from CMA or targeted sequencing. We have diagnosed some rare genetic syndromes using whole-exome sequencing; these diagnoses helped us to better manage the pregnancies.

These choices are part of the case-specific, stepwise approach to genetic evaluation that we take in our fetal heart program. Our goal is to pursue information that will be accurate and valuable for the patient and clinicians, in the most cost-effective and timely manner.
Limitations of noninvasive screening
In our fetal heart program we see increasing numbers of referred patients who have chosen noninvasive cell-free fetal DNA screening (cfDNA) after a cardiac anomaly is detected on ultrasound examination, and who believe that their “low risk” results demonstrate very little or no risk of CHD. Many of these patients express a belief that noninvasive testing is highly sensitive and accurate for fetal anomalies, including CHDs, and are not easily convinced of the value of other genetic tests.
We recently conducted a retrospective chart analysis (unpublished) in which we found that 41% of cases of CHD with abnormal genetics results were not detectable by cfDNA screening.
In the case of atrial-ventricular septal defects and conotruncal abnormalities that often are more associated with common aneuploidies (trisomy 21, 18, 13, and 45 XO), a “high-risk” result from cfDNA screening may offer the family and cardiology/neonatal team some guidance, but a “low-risk” result does not eliminate the risk of a microarray abnormality and thus may provide false reassurance.
Other research has shown that noninvasive screening will miss up to 7.3% of karyotype abnormalities in pregnancies at high risk for common aneuploidies.6
While invasive testing poses a very small risk of miscarriage, it is hard without such testing to elucidate the potential genetic etiologies of CHDs and truly understand the problems. We must take time to thoughtfully counsel patients who decline invasive testing about the limitations of cfDNA screening for CHDs and other anomalies.
Dr. Turan is an associate professor of obstetrics, gynecology, and reproductive sciences, and director of the fetal heart program at the University of Maryland School of Medicine and director of the Fetal Heart Program at the University of Maryland Medical Center. Dr. Turan reported that she has no disclosures relevant to this Master Class. Email her at obnews@mdedge.com.
References
1. J Am Coll Cardiol. 1988 Oct;12(4):1079-86.
2. Pediatr Cardiol. 2019 Mar;40(3):489-96.
3. Ann Pediatr Cardiol. 2017 May-Aug;10(2):126-30.
4. Eur J Obstet Gynecol Reprod Biol 2018;221:172-76.
5. Ultrasound Obstet Gynecol. 2018 Aug;52(2):205-11.
6. PLoS One. 2016 Jan 15;11(1):e0146794.
Congenital heart defects (CHDs) are etiologically heterogeneous, but in recent years it has become clear that genetics plays a larger role in the development of CHDs than was previously thought. Research has been shifting from a focus on risk – estimating the magnitude of increased risk, for instance, based on maternal or familial risk factors – to a focus on the etiology of cardiac defects.
In practice, advances in genetic testing technologies have made the underlying causes of CHDs increasingly detectable. Chromosomal microarray analysis (CMA) – technology that detects significantly more and smaller changes in the amount of chromosomal material than traditional karyotype – has been proven to increase the diagnostic yield in cases of isolated CHDs and CHDs with extracardiac anomalies. Targeted next-generation sequencing also is now available as an additional approach in selective cases, and a clinically viable option for whole-exome sequencing is fast approaching.
For researchers, genetic evaluation carries the potential to unravel remaining mysteries about underlying causes of CHDs – to provide pathological insights and identify potential therapeutic targets. Currently, about 6 % of the total pie of presumed genetic determinants of CHDs is attributed to chromosomal anomalies, 10% to copy number variants, and 12% to single-gene defects. The remaining 72% of etiology, approximately, is undetermined.
As Helen Taussig, MD, (known as the founder of pediatric cardiology) once said, common cardiac malformations occurring in otherwise “normal” individuals “must be genetic in origin.”1 Greater use of genetic testing – and in particular, of whole-exome sequencing – will drive down this “undetermined” piece of the genetics pie.
For clinicians and patients, prenatal genetic evaluation can inform clinical management, guiding decisions on the mode, timing, and location of delivery. Genetic assessments help guide the neonatal health care team in taking optimal care of the infant, and the surgeon in preparing for neonatal surgeries and postsurgical complications.
In a recent analysis of the Society of Thoracic Surgeons Congenital Heart Surgery Database, prenatal diagnosis was associated with a lower overall prevalence of major preoperative risk factors for cardiac surgery.2 Surgical outcomes themselves also have been shown to be better after the prenatal diagnosis of complex CHDs, mainly because of improvements in perioperative care.3
When genetic etiology is elucidated, the cardiologist also is better able to counsel patients about anticipated challenges – such as the propensity, with certain genetic variants of CHD, to develop neurodevelopmental delays or other cardiac complications – and to target patient follow-up. Patients also can make informed decisions about termination or continuation of a current pregnancy and about family planning in the future.
Fortunately, advances in genetics technology have paralleled technological advancements in ultrasound. As I discussed in part one of this two-part Master Class series, it is now possible to detect many major CHDs well before 16 weeks’ gestation. Checking the structure of the fetal heart at the first-trimester screening and sonography (11-14 weeks of gestation) offers the opportunity for early genetic assessment, counseling, and planning when anomalies are detected.
A personalized approach
There has been growing interest in recent years in CMA for the prenatal genetic workup of CHDs. Microarray targets chromosomal regions at a much higher resolution than traditional karyotype. Traditional karyotype assesses both changes in chromosome number as well as more subtle structural changes such as chromosomal deletions and duplications. CMA finds what traditional karyotype identifies, but in addition, it identifies much smaller, clinically relevant chromosomal deletions and duplications that are not detected by karyotype performed with or without fluorescence in-situ hybridization (FISH). FISH uses DNA probes that carry fluorescent tags to detect chromosomal DNA.

At our center, we studied the prenatal genetic test results of 145 fetuses diagnosed with CHDs. Each case involved FISH for aneuploidy/karyotype, followed by CMA in cases of a negative karyotype result. CMA increased the diagnostic yield in cases of CHD by 19.8% overall – 17.4% in cases of isolated CHD and 24.5% in cases of CHD plus extracardiac anomalies.4
Indeed, although a microarray costs more and takes an additional 2 weeks to run, CMA should be strongly considered as first-line testing for the prenatal genetic evaluation of fetuses with major structural cardiac abnormalities detected by ultrasound. However, there still are cases in which a karyotype might be sufficient. For instance, if I see that a fetus has an atrial-ventricular septal defect on a prenatal ultrasound, and there are markers for trisomy 21, 13, or 18, or Turner’s syndrome (45 XO), I usually recommend a karyotype or FISH rather than an initial CMA. If the karyotype is abnormal – which is likely in such a scenario – there isn’t a need for more extensive testing.
Similarly, when there is high suspicion for DiGeorge syndrome (the 22q11.2 deletion, which often includes cleft palate and aortic arch abnormalities), usually it is most appropriate to perform a FISH test.
CMA is the preferred first modality, however, when prenatal imaging suggests severe CHD – for instance, when there are signs of hypoplastic left heart syndrome or tetralogy of Fallot (a conotruncal defect) – or complex CHD with extracardiac anomalies. In these cases, there is a high likelihood of detecting a small deletion or duplication that would be missed with karyotype.
In the past decade, karyotype and CMA have become the major methods used in our practice. However, targeted next‐generation sequencing and whole‐exome sequencing may become more widely used because these technologies enable rapid analysis of a large number of gene sequences and facilitate discovery of novel causative genes in many genetic diseases that cause CHDs.
Currently, targeted next-generation sequencing has mainly been used in the postnatal setting, and there are limited data available on its prenatal use. Compared with whole-exome sequencing, which sequences all of the protein-coding regions of the genome, targeted next-generation sequencing panels select regions of genes that are known to be associated with diseases of interest.
For CHDs, some perinatal centers have begun using a customized gene panel that targets 77 CHD-associated genes. This particular panel has been shown to be useful in addition to current methods and is an effective tool for prenatal genetic diagnosis.5
Whole-exome sequencing is currently expensive and time consuming. While sometimes it is used in the postnatal context, it is not yet part of routine practice as a prenatal diagnostic tool. As technology advances this will change – early in the next decade, I believe. For now, whole-exome sequencing may be an option for some patients who want to know more when severe CHD is evident on ultrasound and there are negative results from CMA or targeted sequencing. We have diagnosed some rare genetic syndromes using whole-exome sequencing; these diagnoses helped us to better manage the pregnancies.

These choices are part of the case-specific, stepwise approach to genetic evaluation that we take in our fetal heart program. Our goal is to pursue information that will be accurate and valuable for the patient and clinicians, in the most cost-effective and timely manner.
Limitations of noninvasive screening
In our fetal heart program we see increasing numbers of referred patients who have chosen noninvasive cell-free fetal DNA screening (cfDNA) after a cardiac anomaly is detected on ultrasound examination, and who believe that their “low risk” results demonstrate very little or no risk of CHD. Many of these patients express a belief that noninvasive testing is highly sensitive and accurate for fetal anomalies, including CHDs, and are not easily convinced of the value of other genetic tests.
We recently conducted a retrospective chart analysis (unpublished) in which we found that 41% of cases of CHD with abnormal genetics results were not detectable by cfDNA screening.
In the case of atrial-ventricular septal defects and conotruncal abnormalities that often are more associated with common aneuploidies (trisomy 21, 18, 13, and 45 XO), a “high-risk” result from cfDNA screening may offer the family and cardiology/neonatal team some guidance, but a “low-risk” result does not eliminate the risk of a microarray abnormality and thus may provide false reassurance.
Other research has shown that noninvasive screening will miss up to 7.3% of karyotype abnormalities in pregnancies at high risk for common aneuploidies.6
While invasive testing poses a very small risk of miscarriage, it is hard without such testing to elucidate the potential genetic etiologies of CHDs and truly understand the problems. We must take time to thoughtfully counsel patients who decline invasive testing about the limitations of cfDNA screening for CHDs and other anomalies.
Dr. Turan is an associate professor of obstetrics, gynecology, and reproductive sciences, and director of the fetal heart program at the University of Maryland School of Medicine and director of the Fetal Heart Program at the University of Maryland Medical Center. Dr. Turan reported that she has no disclosures relevant to this Master Class. Email her at obnews@mdedge.com.
References
1. J Am Coll Cardiol. 1988 Oct;12(4):1079-86.
2. Pediatr Cardiol. 2019 Mar;40(3):489-96.
3. Ann Pediatr Cardiol. 2017 May-Aug;10(2):126-30.
4. Eur J Obstet Gynecol Reprod Biol 2018;221:172-76.
5. Ultrasound Obstet Gynecol. 2018 Aug;52(2):205-11.
6. PLoS One. 2016 Jan 15;11(1):e0146794.